|
|
|
|
Application of structure to data from the Taita thrush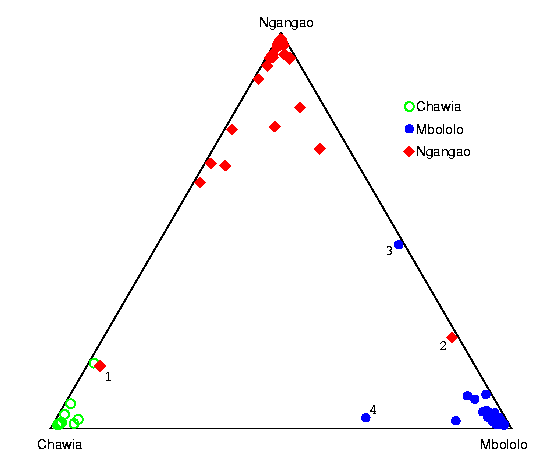
155 individuals of the Kenyan thrush ( T. helleri) were collected in three separate refugia of indigenous cloud forest (Chawia, Mbololo, Ngangao); each individual was genotyped at seven microsatellite loci (Galbusera et al, 2000, Conservation Genetics 1:45). The original study was aimed to address questions including: Do these three refugia represent genetically distinct subpopulations? Is there evidence for recent migration between refugia? The plot summarizes our results for one analysis of these data. Here, we estimated the ancestry of individuals under a model in which there are three subpopulations, and individuals can have mixed ancestry (the model with three populations has by far the highest posterior support). Each point shows an estimate of the amount of ancestry that an individual has in each of the three subpopulations (given by the distances to each side of the triangle). The points are labelled according to sampling location, but we were not told the locations until after obtaining these results. Four individuals who have moderate posterior support for having migrant parents are labelled '1'--'4'. A Neighbor-Joining tree of these data is relatively uninformative. Our results are presented in detail in Pritchard et al 2000. [PDF] I have prepared a sequence of plots illustrating the convergence of the algorithm from a random initial state. Data courtesy of Dr. Peter Galbusera.
|