



Next: Specifying initial and boundary
Up: Creating or Reading in
Previous: Using the triangle libraries
Contents
Reading triangular grids from a file
Use of the -t flag creates three files specified in suntans.dat:
points, cells, and edges. By default, these are specified
to be
points points.dat
edges edges.dat
cells cells.dat
The points file contains a listing of the x-y coordinates of the Delaunay points
in the full triangulation before being subdivided among different processors.
This file contains three columns although the last column is never used. The
total number of lines in this file is 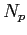 , the number of triangle vertics in the triangulation.
The
, the number of triangle vertics in the triangulation.
The edges file contains  rows each of which defines an
edge in the triangulation, and five columns in the following format:
rows each of which defines an
edge in the triangulation, and five columns in the following format:
Point1 Point2 Marker Voronoi1 Voronoi2
Point1 and Point2 contain indices to points in the points file
and make up the end points of the Delaunay edges. Because SUNTANS uses C-style
indexing, then 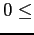
Point1,Point2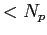 .
. Marker specifies the type
of edge. If Marker=0, then it is a computational edge, otherwise, it is
a boundary edge, and the boundary condition is specified in Section 4.2.
The last two entries, Voronoi1 and Voronoi2, are the indices to the Voronoi
points which make up the end points of the Voronoi edge which intersects this Delaunay edge.
As such, we must have 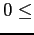
Voronoi1,Voronoi2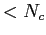 .
These Voronoi points correspond to triangles defined in the file
.
These Voronoi points correspond to triangles defined in the file cells. Voronoi points
which are ghost points are indicated by a 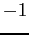 .
The
.
The cells file contains  rows each of which corresponds to a
triangle in the triangulation, and 8 columns in the following format:
rows each of which corresponds to a
triangle in the triangulation, and 8 columns in the following format:
xv yv Point1 Point2 Point3 Neigh1 Neigh2 Neigh3
The xv and yv points correspond to the x-y coordinates
of the Voronoi points of each triangle and Point1, Point2, and Point3
correspond to indices to points in the points file which make up the
vertices of the triangle. These indices must satisfy
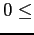
Point1,Point2,Point3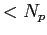 .
.
Neigh1, Neigh2, and
Neigh3 correspond to indices to neighboring triangles. Neighboring
triangles which correspond to ghost points are represented by a 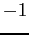 . For neighbors
not lying outside boundaries, we must have
. For neighbors
not lying outside boundaries, we must have
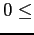
Neigh1,Neigh2,Neigh3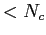 .
.
Because SUNTANS determines the number of triangle vertices 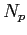 , edges
, edges  , and cells
, and cells  by the number of rows in the
by the number of rows in the points, cells, and edges files, respectively,
it is important not to have extra carriage returns at the end of these files.
These three files are generated each time the -t flag is used with SUNTANS. If
the -t flag is not used, then when called with 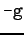 , SUNTANS reads these
three files and computes grid geometry and, if desired, partitions it among several processors.
The
, SUNTANS reads these
three files and computes grid geometry and, if desired, partitions it among several processors.
The -g flag outputs the following data files, which are specified in suntans.dat.
One file associated with each of these descriptors is created for each processor in a partitioned
grid. For example, if the file name specified after cells in suntans.dat is given
by cells.dat, then when called with 2 processors, the -g flag would output two files
names cells.dat.0 and cells.dat.1, each corresponding to the cells file of
each processor.
cells Same as the output when using -t, except on a per-processor basis.
The indices to the triangle vertices still correspond to indices in the global points file, which is
not distributed among the processors. All other indices are local to the specific processor.
edges Same as the output when using -t, except on a per-processor basis.
The indices to the end points of the edges still correspond to indices in the global points file, which is
not distributed among the processors. All other indices are local to the specific processor.
celldata Contains the grid data associated with the Voronoi points of each cell and contains
 rows, where
rows, where  is the number of cells on each processor (including interprocessor ghost points).
Each row contains the following entries:
is the number of cells on each processor (including interprocessor ghost points).
Each row contains the following entries:
xv yv Ac dv Nk Edge{1-3} Neigh{1-3} N{1-3} def{1-3}
xv yv are the Voronoi coordinates
Ac is the cell area
dv is the depth at the point xv,yv. This is the depth of the bottom-most
face of the column beneath this cell and is not the actual depth, which is always greater than
dv.
Nk is the number of vertical levels in the water column.
Edge{1-3} are indices to the three edges that correspond to the faces of the cell.
Neigh{1-3} are the indices to the three neighboring cells.
N{1-3} is the dot product of the unique normal with the outward normal on each
face.
def{1-3} is the distance from the Voronoi point to the three faces.
edgedata Contains the grid data associated with the Delaunay edges and contains
 rows, where
rows, where  is the number of edges on each processor (including interprocessor edges).
Each row contains the following entries:
is the number of edges on each processor (including interprocessor edges).
Each row contains the following entries:
df dg n1 n2 xe ye Nke Nkc grad{1,2} gradf{1,2} mark xi{1,2,3,4} eneigh{1,2,3,4}
df is the length of the edge.
dg is the distance between the Voronoi points on either side of the edge. If
this is a boundary edge then dg is twice the distance between the edge and the Voronoi
point on the inside of the boundary.
n1,n2 are the components of the normal direction of the edge. These correspond
to the unique normals of each edge. The outward normal for this edge corresponding to
a particular cell is given by n1*N, n2*N, where N is the dot product of the
unique normal with the outward normal and is specified in the celldata file.
xe, ye are the coordinates of the intersection of the edge with the Delaunay
edge.
Nke is the number of active edges in the vertical (see Nkc).
Nkc is the maximum number of active cells in the vertical which neighbor a given edge. Nkc is
always at least Nke. See Figure 1 for a graphical depiction.
Figure 1:
Depiction of an edge in which Nke=4 and Nkc=6.
.5 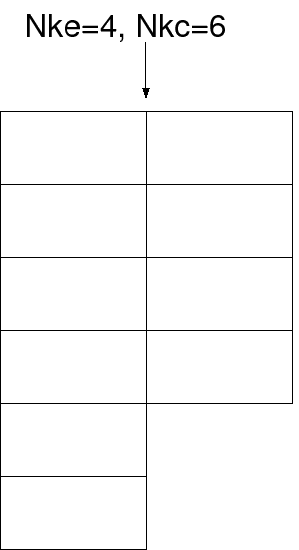
|
item grad{1,2} are indices to the Voronoi points defined in the celldata file.
If  is the number of cells on a processor, then
is the number of cells on a processor, then 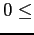
grad{1,2}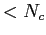 .
.
gradf{1,2} are indices that determine the location of the edge in the ordering
of the Edge{1-3} or def{1-3} arrays. Each cell contains a pointer to this edge
in its list of Edge{1-3} pointers. The gradf{1,2} index is a number from
0 to 2 which determines which face number this edge is of a particular cell.
mark Contains the marker type for this edge. All edges with the value 0 are
computational edges, while other values are described in Section 4.2.
topology
Np Nneighs neighbor{0,1,2,...,Np-1}\n
neigh0: num_cells_send num_cells_recv num_edges_send num_edges_recv
cell_send_indices ...
cell_receive_indices ...
edge_send_indices ...
edge_recv_indices ...
neigh1: num_cells_send num_cells_recv num_edges_send num_edges_recv
cell_send_indices ...
cell_receive_indices ...
edge_send_indices ...
edge_recv_indices ...
.
.
.
neigh{Numneighs-1}: num_cells_send num_cells_recv num_edges_send num_edges_recv
cell_send_indices ...
cell_receive_indices ...
edge_send_indices ...
edge_recv_indices ...
celldist[0] celldist[1] celldist[2] ... celldist[MAXBCTYPES-1]
edgedist[0] edgedist[1] edgedist[2] ... edgedist[MAXBCTYPES-1]
cellp[0],...,cellp[Nc-1]
edgep[0],...,edgep[Ne-1]
vertspace Contains the vertical grid spacings. This file has Nkmax rows,
where Nkmax is the number of z-levels.




Next: Specifying initial and boundary
Up: Creating or Reading in
Previous: Using the triangle libraries
Contents
2014-08-06
