Polyketide Synthases (PKSs)

Assembly-line enzymes such as polyketide synthases (PKSs) have extraordinary potential for the programmable biosynthesis of complex natural products. Our laboratory seeks to understand the mechanistic logic of assembly-line PKSs, and to harness these insights in order to engineer new antibiotics. The prototypical system of interest to us is the 6-deoxyerythronolide B synthase (DEBS), which synthesizes the macrocyclic core of the antibiotic erythromycin. We are interested in elements such as acyltransferase specificity, module/domain engineering, and structure-function relationships. Additionally, we hope to elucidate the structure and biological mode-of-action of orphan polyketide products.
Understanding the Catalytic Cycles of the 6-Deoxyerythronolide B Synthase (DEBS)

We can use monoclonal Fabs to trap a conformation during specific stages of the catalytic cycle to understand the reactions carried out by DEBS.
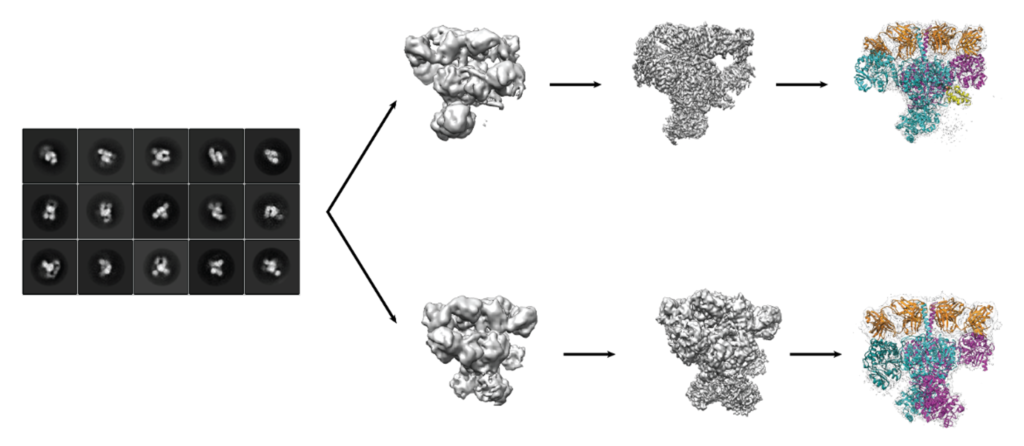
We are using single-particle cryo-EM to understand the various conformations that exist during the catalytic cycle.
De-Orphanizing the NOCardiosis-Associated Polyketide (NOCAP)

Recombinant protein expression for in vitro biosynthesis and analysis paired with heterologous biosynthesis in E. coli elucidated the chemical structure of the NOCAP aglycone product. We soon hope to characterize the fully decorated natural product to understand its biological activity.
Uncovering the relationship between the NOCAP Synthase & Nocardiosis
We are developing NOCAP synthase (+) and (-) nocardia strains for future human cell line infection assays.
Evolution of Assembly-Line PKSs and Diversification of their Natural Products
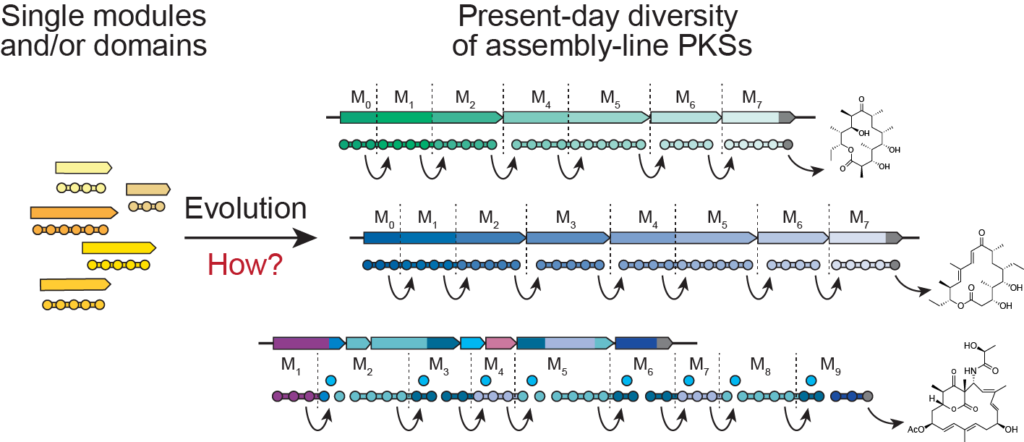
By studying a newly discovered genetic element present in most assembly-line PKSs, we investigate the mechanism by which multi-modularity in PKSs evolved. We are looking to harness evolutionary strategies to generate chemical diversity in the lab.
Celiac Disease
Celiac disease is a T cell driven autoimmune disease of the small intestine that is induced by exposure to gluten from foodgrains such as wheat, rye and barley. Our laboratory seeks to understand the earliest molecular recognition and catalytic events in the pathogenic response of the celiac intestine to dietary gluten. We anticipate that such insights will pave the way for new therapies and biomarkers for this widespread but overlooked disorder. Our present efforts are directed at testing the hypothesis that transglutaminase 2 (TG2), the principal autoantigen associated with celiac disease, is also a druggable target for therapeutic intervention.
Developing in vivo compatible probes of active TG2 for the study of celiac disease pathogenesis
LRP-1 mediated antigen endocytosis and presentation pathway
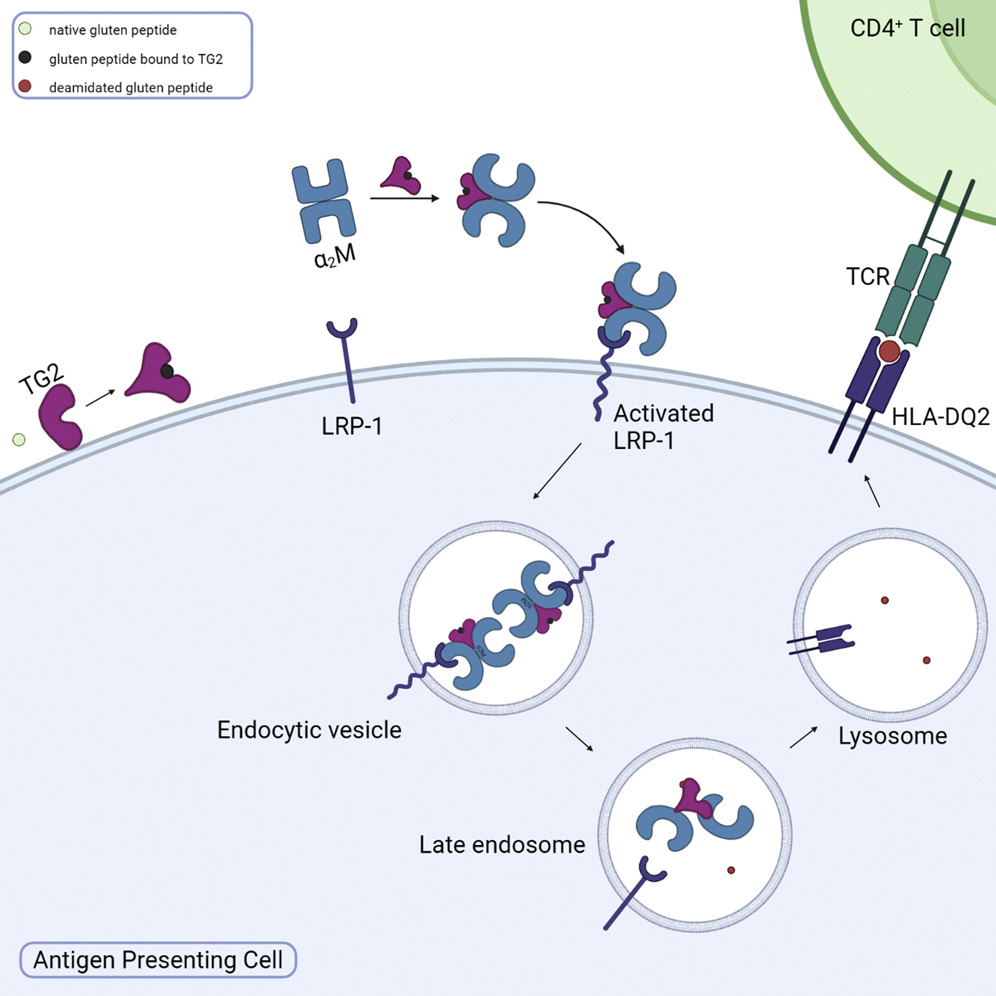
Using novel chemical probes for active TG2 we have described a pathway through which gluten peptides, the primary celiac disease antigen is endocytosed, processed and presented on MHC-II. We are now seeking to understand the effect of this pathway on downstream T cell activation and inflammation. We are also interested in pinpointing the subsets of cells in the gut that are important for antigen presentation through this pathway. Finally, we are developing chemical tools to take advantage of this efficient, TG2/gluten complex specific endocytosis pathway.
Characterization of TG2 with Substrates and Protein Partners
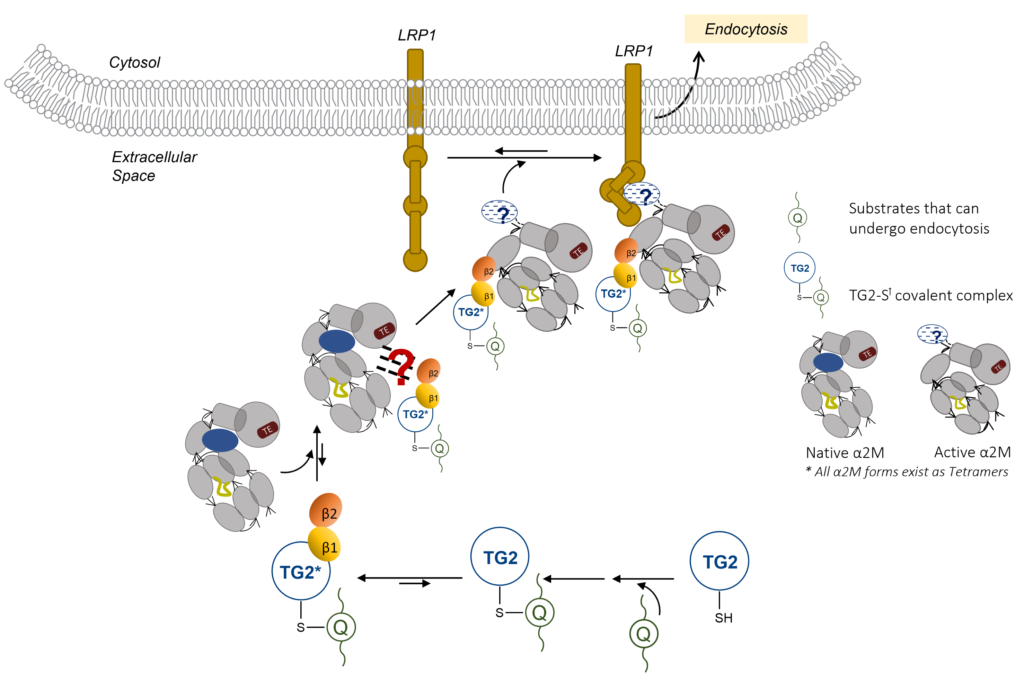
LRP1 endocytosis of TG2-Gluten Complex needs β-Barrel Recognition by α2-Macroglobulin. To further characterize the interactions in this pathway, we are have adopted structural (X-Ray Crystallography, SEC-SAXS) and biochemical analysis that measures binding of this four-component system. When combined, these studies may give insight into the selective endocytosis via the LRP1 Pathway.
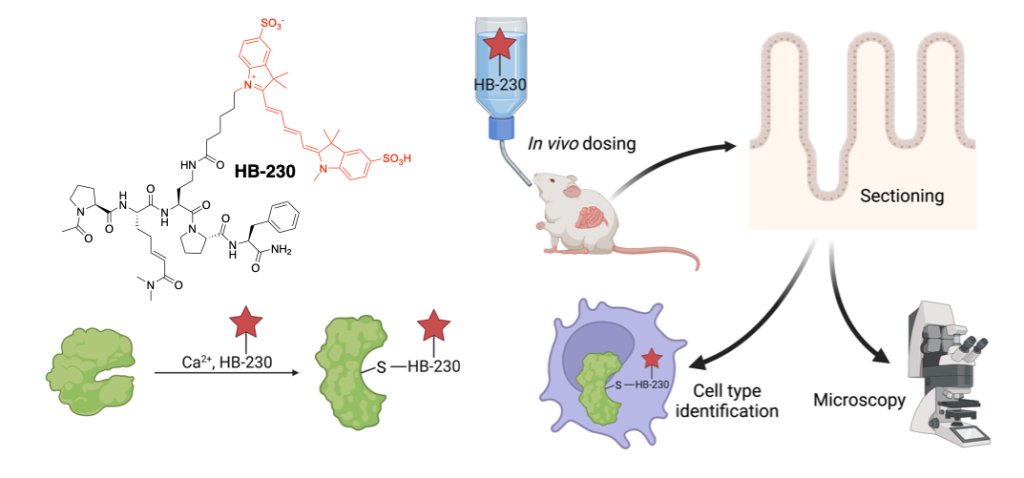
PK/PD/mouse studies of HB230
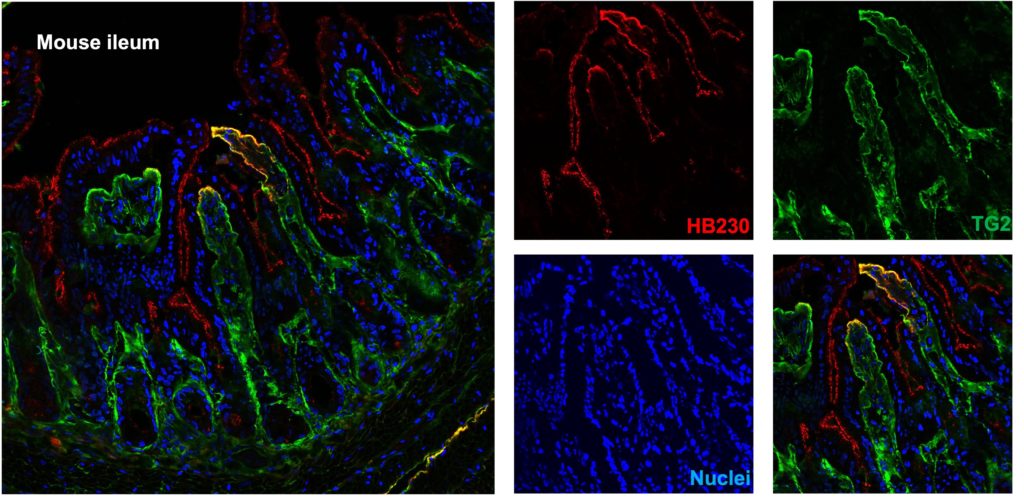
Using a fluorescent-tagged chemical probe (HB-230) to track active TG2 distribution in the mouse small intestine in vivo. Here we identified HB-230 in the lamina propria of mouse ileum, which is colocalized with TG2, demonstrating that TG2/gluten complex uptake and presentation by immune cells activate T cells in the small intestine.