- ... Pritchard1
- Our other colleagues in the structure project are Peter Donnelly, Daniel Falush and Matthew Stephens.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...12
- The first version of this program was developed while the authors (JP, MS, PD) were in the Department of Statistics, University of Oxford.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...23
- Comments and queries can be emailed to us at structure_help@genetics.uchicago.edu. In view of the considerable volume of email that we receive, please check this document carefully first.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... program4
- The
software and the manuscript are available from http://pritch.bsd.uchicago.edu. (Click on ``software''.)
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
chunk.5
- Because of the way that this is parameterized, the map
distances in the input file can be in arbitrary units-e.g., genetic
distances, or physical distances (under the assumption that these are
roughly proportional to genetic distances). Then the estimated value
of
 represents the rate of switching from one chunks to the next,
per unit of whatever distance was assumed in the input file. E.g., if
an admixture event took place ten generations ago, then
represents the rate of switching from one chunks to the next,
per unit of whatever distance was assumed in the input file. E.g., if
an admixture event took place ten generations ago, then  should be
estimated as
should be
estimated as 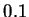 when the map distances are measured in cM (this is
when the map distances are measured in cM (this is
 , where 0.01 is the probability of recombination per
centiMorgan), or as
, where 0.01 is the probability of recombination per
centiMorgan), or as
 when the map distances are
measured in KB (assuming a constant crossing-over rate of 1cM/MB).
The prior for
when the map distances are
measured in KB (assuming a constant crossing-over rate of 1cM/MB).
The prior for  is log-uniform. The front end tries to make some
guesses about sensible upper and lower bounds for
is log-uniform. The front end tries to make some
guesses about sensible upper and lower bounds for  , but the user
should adjust these to match the biology of the situation.
, but the user
should adjust these to match the biology of the situation.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... off.6
- If the admixture model is used to
estimate
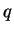 for those individuals without prior population
information,
for those individuals without prior population
information, 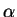 is updated on the basis of those individuals
only. If there are very few such individuals, you may need to fix
is updated on the basis of those individuals
only. If there are very few such individuals, you may need to fix
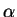 at a sensible value.
at a sensible value.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... below.7
- The model of
correlated allele frequencies implemented now differs slightly from
that in structure Version 1.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... modes8
- For
 here are
always multiple symmetric modes that correspond to permuting the
labels; these do not cause serious problems Pritchard et al. (2000a)
here are
always multiple symmetric modes that correspond to permuting the
labels; these do not cause serious problems Pritchard et al. (2000a)
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.