



Next: 3. Linkage model
Up: Ancestry Models
Previous: 1. No admixture model
Contents
Individuals may have mixed ancestry. This is modelled by saying that
individual 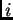 has inherited some fraction of his/her genome from
ancestors in population
has inherited some fraction of his/her genome from
ancestors in population 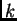 . The output records the posterior
mean estimates of these proportions. Conditional on the ancestry
vector,
. The output records the posterior
mean estimates of these proportions. Conditional on the ancestry
vector, 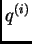 , the origin of each allele is independent.
We recommend this model as a starting point for most analyses. It is
a reasonably flexible model for dealing with many of the complexities
of real populations. Admixture is a common feature of real data, and
you probably won't find it if you use the no-admixture model. The
admixture model can also deal with hybrid zones in a natural way.
, the origin of each allele is independent.
We recommend this model as a starting point for most analyses. It is
a reasonably flexible model for dealing with many of the complexities
of real populations. Admixture is a common feature of real data, and
you probably won't find it if you use the no-admixture model. The
admixture model can also deal with hybrid zones in a natural way.
William Wen
2002-07-18