



Next: 4. Using prior population
Up: Ancestry Models
Previous: 2. Admixture model
Contents
This is essentially a generalization of the admixture model to deal
with ``admixture linkage disequilibrium''-i.e., the correlations that
arise between linked markers in recently admixed populations. There
is a manuscript in preparation (Falush, Stephens and Pritchard) that
describes the model, and computations in more detail.
The basic model is that, 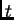 generations in the past, there was an
admixture event that mixed the
generations in the past, there was an
admixture event that mixed the 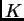 populations. If you consider an
individual chromosome, it is composed of a series of ``chunks'' that
are inherited as discrete units from ancestors at the time of the
admixture. Admixture LD arises because linked alleles are often on
the same chunk, and therefore come from the same ancestral population.
The sizes of the chunks are assumed to be independent exponential
random variables with mean length
populations. If you consider an
individual chromosome, it is composed of a series of ``chunks'' that
are inherited as discrete units from ancestors at the time of the
admixture. Admixture LD arises because linked alleles are often on
the same chunk, and therefore come from the same ancestral population.
The sizes of the chunks are assumed to be independent exponential
random variables with mean length 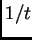 (in Morgans). In practice we
estimate a ``recombination rate''
(in Morgans). In practice we
estimate a ``recombination rate'' 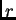 from the data that corresponds
to the rate of switching from the present chunk to a new
chunk.5 Each
chunk in individual
from the data that corresponds
to the rate of switching from the present chunk to a new
chunk.5 Each
chunk in individual 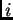 is derived independently from population
is derived independently from population
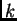 with probability
with probability 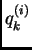 , where
, where 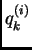 is the proportion
of that individual's ancestry from population
is the proportion
of that individual's ancestry from population 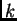 .
Overall, the new model retains the main elements of the admixture
model, but all the alleles that are on a single chunk have to come
from the same population. The new MCMC algorithm integrates over the
possible chunk sizes and break points. It reports the overall
ancestry for each individual, taking account of the linkage, and can
also report the probability of origin of each bit of chromosome, if
desired by the user.
This new model performs better than the original admixture model when
using linked loci to study admixed populations. It achieves more
accurate estimates of the ancestry vector, and can extract more
information from the data. It should be useful for admixture mapping.
Clearly, this model is a big simplification of the complex realities
of most real admixed populations. However, the major effect of admixture
is to create long-range correlation among linked markers, and so our
aim here is to encapsulate that feature within a fairly simple model.
The computations are a bit slower than for the admixture model,
especially with large
.
Overall, the new model retains the main elements of the admixture
model, but all the alleles that are on a single chunk have to come
from the same population. The new MCMC algorithm integrates over the
possible chunk sizes and break points. It reports the overall
ancestry for each individual, taking account of the linkage, and can
also report the probability of origin of each bit of chromosome, if
desired by the user.
This new model performs better than the original admixture model when
using linked loci to study admixed populations. It achieves more
accurate estimates of the ancestry vector, and can extract more
information from the data. It should be useful for admixture mapping.
Clearly, this model is a big simplification of the complex realities
of most real admixed populations. However, the major effect of admixture
is to create long-range correlation among linked markers, and so our
aim here is to encapsulate that feature within a fairly simple model.
The computations are a bit slower than for the admixture model,
especially with large 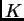 and unphased data. Nonetheless, they are
practical for hundreds of sites and individuals and multiple
populations. The model can only be used if there is information about
the relative positions of the markers (usually a genetic map).
and unphased data. Nonetheless, they are
practical for hundreds of sites and individuals and multiple
populations. The model can only be used if there is information about
the relative positions of the markers (usually a genetic map).




Next: 4. Using prior population
Up: Ancestry Models
Previous: 2. Admixture model
Contents
William Wen
2002-07-18