


Next:Allele
frequency modelsUp:Ancestry
ModelsPrevious:3.
Linkage modelContents
4. Using prior population information
The default mode for structure uses only genetic information to
learn about population structure. However, there is often other information
that can be used (e.g., physical characteristics of sampled individuals
or geographic sampling location) to assist the clustering. In the section
``Model with prior population information'' in (Pritchard
et al. 2000a), we described our framework
for incorporating this type of information into the inference procedure.
There are at least two kinds of reasons for making use of this sort of
extrinsic population information. One is that one may want to test whether
any individuals in the sample are immigrants to their supposed populations,
or have recent immigrant ancestors (see the example in Pritchard
et al. 2000a). A second is that we may want to make use of learning
samples: ie., we have some individuals of known origin, and we want to
use them to help us classify individuals of unknown origin. For example
in
Beaumont et al., 2001, we wanted
to learn about the ancestry of Scottish wildcats (many of which are hybridized
with feral domestic cats). We had genetic data from a bunch of pet house
cats which we defined as being in one population, while we inferred 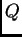 for the wildcats (with
for the wildcats (with 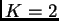 ).
Use of this sort of prior information will normally improve the accuracy
of the inference. To use these options you need to set USEPOPINFO to 1,
and choose a value of MIGRPRIOR (which is
).
Use of this sort of prior information will normally improve the accuracy
of the inference. To use these options you need to set USEPOPINFO to 1,
and choose a value of MIGRPRIOR (which is 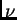 in Pritchard et al). You might choose something in the range 0.001
to 0.1 for
in Pritchard et al). You might choose something in the range 0.001
to 0.1 for 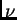 .
Even when using learning samples, it may be sensible to allow for some
misclassification by setting MIGRPRIOR larger than 0. The pre-defined population
for each individual is set in the input data file (see PopData) and should
be an integer between 1 and MAXPOPS, inclusive. If PopData for any individual
is outside this range, their
.
Even when using learning samples, it may be sensible to allow for some
misclassification by setting MIGRPRIOR larger than 0. The pre-defined population
for each individual is set in the input data file (see PopData) and should
be an integer between 1 and MAXPOPS, inclusive. If PopData for any individual
is outside this range, their 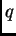 will be updated in the normal way (ie without prior population information,
according to the model that would be used if USEPOPINFO was turned off.6).
Learning samples are implemented through the use of the PopFlag column
in the data file. The pre-defined population is used for those individuals
for whom PopFlag=1, and it is ignored for individuals for whom PopFlag=0.
If there is no PopFlag column in the data file, then when USEPOPINFO is
turned on, PopFlag is set to 1 for all individuals. In general, we advocate
that the user should first run the program without population information
to ensure that the pre-defined populations are in rough agreement with
the genetic information.
will be updated in the normal way (ie without prior population information,
according to the model that would be used if USEPOPINFO was turned off.6).
Learning samples are implemented through the use of the PopFlag column
in the data file. The pre-defined population is used for those individuals
for whom PopFlag=1, and it is ignored for individuals for whom PopFlag=0.
If there is no PopFlag column in the data file, then when USEPOPINFO is
turned on, PopFlag is set to 1 for all individuals. In general, we advocate
that the user should first run the program without population information
to ensure that the pre-defined populations are in rough agreement with
the genetic information.



Next:Allele
frequency modelsUp:Ancestry
ModelsPrevious:3.
Linkage modelContents
William Wen 2002-07-18