| (1) |
| (1) |
independently for each ![]() .
Then the prior for the frequencies in population
.
Then the prior for the frequencies in population ![]() is
is
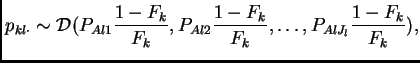 |
(2) |
independently for each ![]() and
and ![]() .
In this model, the
.
In this model, the ![]() s
have a close relationship to the standard measure of genetic distance,
s
have a close relationship to the standard measure of genetic distance, ![]() .
In the standard parametrization of
.
In the standard parametrization of ![]() ,
the expected frequency in each population is given by overall mean frequency,
and the variance in frequency across subpopulations of an allele at overall
frequency
,
the expected frequency in each population is given by overall mean frequency,
and the variance in frequency across subpopulations of an allele at overall
frequency![]() is
is ![]() .
The model here is much the same, except that we generalize the model slightly
by allowing each population to drift away from the ancestral population
at a different rate (
.
The model here is much the same, except that we generalize the model slightly
by allowing each population to drift away from the ancestral population
at a different rate (![]() ),
as might be expected if populations have different sizes. We also try to
estimate ``ancestral frequencies'', rather than using the mean frequencies.
We have placed independent priors on the
),
as might be expected if populations have different sizes. We also try to
estimate ``ancestral frequencies'', rather than using the mean frequencies.
We have placed independent priors on the ![]() ,
proportional to a gamma distribution with means of 0.01 and standard deviation
0.05 (but with
,
proportional to a gamma distribution with means of 0.01 and standard deviation
0.05 (but with ![]() ).
The parameters of the gamma prior can be modified by the user. Some experimentation
suggests that the prior mean of 0.01, which corresponds to very low levels
of subdivision, often leads to good performance for data that are difficult
for the independent frequencies model. In other problems, where the differences
among populations are more marked, it seems that the data usually overwhelm
this prior on
).
The parameters of the gamma prior can be modified by the user. Some experimentation
suggests that the prior mean of 0.01, which corresponds to very low levels
of subdivision, often leads to good performance for data that are difficult
for the independent frequencies model. In other problems, where the differences
among populations are more marked, it seems that the data usually overwhelm
this prior on ![]() .
.