



Next: Sequence data and Y
Up: Missing data, null alleles...
Previous: Missing data, null alleles...
Contents
For some types of genetic markers, it is not possible to distinguish
all the genotypes (eg, genotypes AA and Aa look the same
on the gel, while aa is different). I have not implemented
anything specific for this type of data, however the program can
be applied to such data under the ``no admixture'' model (set
NOADMIX=1).
This can done by treating each class of genotypes as being,
effectively, a haploid allele. Then we might designate an AA/Aa
individual in the input file as (1,-9), and an aa individual as
(2,-9), where -9 is the value used for missing data. This approach
can also be used for more than two genotypic classes.
This fix is valid under the no-admixture model, because the updates
for 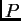 and
and 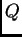 are still correct even though we are treating
genotypic classes rather than alleles. Of course, there is less
information than if all the genotypes could be distinguished. This
fix is not formally correct under the admixture model, however it
seems plausible that admixture estimates may be reasonably unbiased in
this situation too, especially if there are lots of loci. We have not
looked at this.
are still correct even though we are treating
genotypic classes rather than alleles. Of course, there is less
information than if all the genotypes could be distinguished. This
fix is not formally correct under the admixture model, however it
seems plausible that admixture estimates may be reasonably unbiased in
this situation too, especially if there are lots of loci. We have not
looked at this.




Next: Sequence data and Y
Up: Missing data, null alleles...
Previous: Missing data, null alleles...
Contents
William Wen
2002-07-18