


Next: Missing genotype data
Up: Format for the data
Previous: Rows
Contents
Each row of individual data contains the following elements; these
are columns in the data file.
- Label (Optional; string) A string of integers or characters used to
designate each individual in the sample.
- PopData (Optional; integer) An integer designating a user-defined
population from which the individual was obtained (for instance these
might designate the geographic sampling locations of individuals).
- PopFlag (Optional; 0 or 1) A Boolean flag which indicates whether
to use the PopData when using learning learning samples (see
USEPOPINFO, below). (Note: A Boolean variable (flag) is a
variable which takes the values TRUE or FALSE, which are designated
here by the integers 1 (use PopData) and 0 (don't use PopData),
respectively.)
- Phenotype (Optional; integer) An integer designating the value of a
phenotype of interest, for each individual. (
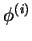 in table.)
(The phenotype information is not actually used in this program. It
is here to permit a smooth interface with the program STRAT which
is used for association mapping.)
in table.)
(The phenotype information is not actually used in this program. It
is here to permit a smooth interface with the program STRAT which
is used for association mapping.)
- Extra Columns (Optional; string) It may be convenient for
the user to include additional data in the input file which are
ignored by the program. These go here, and may be strings of integers
or characters.
- Genotype Data (Required; integer) Each allele at a given
locus should be coded by a unique integer (eg microsatellite repeat
score).



Next: Missing genotype data
Up: Format for the data
Previous: Rows
Contents
Jonathan Pritchard
2003-07-10