


Next: Parameters in file extraparams.
Up: Running structure from the
Previous: Running structure from the
Contents
Parameters in file mainparams.
The user will need to set all of these parameters before running the
program. Several of these parameters (LABEL, POPDATA, POPFLAG,
PHENOTYPE, EXTRACOLS) indicate whether particular types of data are
present in the input file; these are described in Section 3.
- INFILE (string) Name of input data file. Max length 30
characters (or possibly less depending on operating system).
- OUTFILE (string) Name for program output files (the suffixes ``_1'',
``_2'', ...,``_
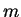 '' (for intermediate results) and ``_f'' (final
results) are added to this name). Existing files with these names
will be overwritten. Max length of name 30 characters (or possibly
less depending on operating system).
'' (for intermediate results) and ``_f'' (final
results) are added to this name). Existing files with these names
will be overwritten. Max length of name 30 characters (or possibly
less depending on operating system).
- NUMINDS (int) Number of diploid individuals in data file.
- NUMLOCI (int) Number of loci in data file.
- LABEL (Boolean) Input file contains labels (names) for each
individual. 1 = Yes; 0 = No.
- POPDATA (Boolean) Input file contains a user-defined
population-of-origin for each individual. 1 = Yes; 0 = No.
- POPFLAG (Boolean) Input file contains an indicator variable which says
whether to use popinfo when USEPOPINFO==1 (see below). 1 = Yes; 0 = No.
- PHENOTYPE (Boolean) Input file contains a column of phenotype
information. 1 = Yes; 0 = No.
- EXTRACOLS (int) Number of additional columns of data after the
Phenotype before the genotype data start. These are ignored by the
program. 0 = no extra columns.
- PHASEINFO (Boolean) The row(s) of genotype data for each
individual are followed by a row of information about haplotype phase.
This is for use with the linkage model only. See sections
3 and 4.2 for further details.
- MISSING (int) Value given to missing genotype data. Must be an
integer, and must not appear elsewhere in the data set. I use -9.
- PLOIDY (int) Ploidy of the organism. Default is 2 (diploid).
- ONEROWPERIND (Boolean) The data for each individual are arranged
in a single row. E.g., for diploid data, this would mean that the two alleles
for each locus are in consecutive order in the same row, rather than being
arranged in the same column, in two consecutive rows. See section
3 for details about input formats.
- GENENAMES (Boolean) The top row of the data file contains a list
of
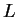 names corresponding to the markers used.
names corresponding to the markers used.
- MAPDISTANCES (Boolean) The next row of the data file (or
the first row if GENENAMES==0) contains a list of mapdistances between
neighboring loci.
- MAXPOPS (int) Number of populations assumed for a particular
run of the program. Pritchard et al. (2000a) call this
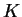 . Sometimes
(depending on the nature of the data) there is a natural value of
. Sometimes
(depending on the nature of the data) there is a natural value of 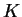 that can be used, otherwise
that can be used, otherwise 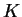 can be estimated by checking the fit
of the model at different values of
can be estimated by checking the fit
of the model at different values of 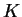 (see Section
5).
(see Section
5).
- BURNIN (int) Length of burnin period before the start of data collection.
(See Section 4.1.)
- NUMREPS (int) Number of MCMC reps after burnin. (See Section 4.1.)



Next: Parameters in file extraparams.
Up: Running structure from the
Previous: Running structure from the
Contents
Jonathan Pritchard
2003-07-10