


Next: Output options
Up: Running structure from the
Previous: Parameters in file mainparams.
Contents
These options allow the user to refine the model in various ways, and
do more involved analyses. The default values are probably fine to
begin with. For Boolean options, type 1 for ``Yes'', or ``Use this
option''; 0 for ``No'' or ``Don't use this option''.
- FREQSCORR (double) Use a model where the allele frequencies
are correlated. More specifically, rather than assuming a prior in
which the allele frequencies in each population are independent draws
from a uniform Dirichlet distribution, we start with a distribution
which is centered around the mean allele frequencies in the sample.
This model is more realistic for very closely related populations
(where we expect the allele frequencies to be similar across
populations), and can produce better clustering (section
4.3). The prior of
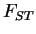 is set using FPRIORMEAN,
and FPRIORSD. There may be a tendency to overestimate
is set using FPRIORMEAN,
and FPRIORSD. There may be a tendency to overestimate 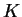 when
FREQSCORR is turned on.
when
FREQSCORR is turned on.
- ONEFST (Boolean) Assume the same value of
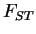 for all
populations. This is not recommended for most data, because in
practice you probably expect different levels of divergence in each
population. The important exception is if you are running the program
for
for all
populations. This is not recommended for most data, because in
practice you probably expect different levels of divergence in each
population. The important exception is if you are running the program
for 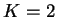 , and you care about the actual value of
, and you care about the actual value of 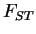 , because in
that case it is not really possible to estimate two values of
, because in
that case it is not really possible to estimate two values of 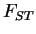 separately. When you're trying to estimate
separately. When you're trying to estimate 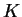 , you should use the
same model for all
, you should use the
same model for all 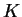 (I'd suggest ONEFST=0).
(I'd suggest ONEFST=0).
- INFERALPHA (Boolean) Infer the value of the
model parameter
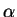 from the data; otherwise
from the data; otherwise 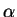 is fixed at
the value ALPHA which is chosen by the user. This option is ignored
under the NOADMIX model. (The prior for the ancestry vector
is fixed at
the value ALPHA which is chosen by the user. This option is ignored
under the NOADMIX model. (The prior for the ancestry vector 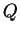 is
Dirichlet with parameters
is
Dirichlet with parameters
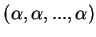 . Small
. Small
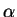 implies that most individuals are essentially from one
population or another, while
implies that most individuals are essentially from one
population or another, while 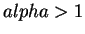 implies that most individuals
are admixed.)
implies that most individuals
are admixed.)
- POPALPHAS (Boolean) Infer a separate
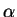 for each
population. Not recommended in most cases but may be useful for
situations with asymmetric admixture.
for each
population. Not recommended in most cases but may be useful for
situations with asymmetric admixture.
- RECOMBINE (Boolean) Use the linkage model. See section
4.2. RLOG10START sets the initial value of
recombination rate r per unit distance. RLOG10MIN and
RLOG10MAX set the minimum and maximum allowed values for
log10r. RLOG10PROPSD sets the size of the proposed changes to log10r
in each update. The front end makes some guesses about these, but some
care on the part of the user in required to be sure that the values
are sensible for the particular application.
- COMPUTEPROB (Boolean) Print the log-likelihood of the data
at each update, and estimate the probability of the data given
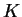 and
the model (see section 5). This is used in estimating
and
the model (see section 5). This is used in estimating
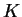 , and is also a useful diagnostic for whether the burnin is long
enough. The main reason for turning this off would be to speed up the
program (
, and is also a useful diagnostic for whether the burnin is long
enough. The main reason for turning this off would be to speed up the
program (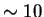 -
-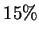 ).
).
- INFERLAMBDA (Boolean) Infer a suitable value for
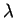 .
Not recommended for most analyses.
.
Not recommended for most analyses.
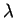 parameterizes the allele frequency prior, and for most data
the default value of 1 seems to work pretty well. If the frequencies
at most markers are very skewed towards low/high frequencies, a
smaller value of
parameterizes the allele frequency prior, and for most data
the default value of 1 seems to work pretty well. If the frequencies
at most markers are very skewed towards low/high frequencies, a
smaller value of 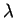 may potentially lead to better performance.
It doesn't seem to work very well to estimate
may potentially lead to better performance.
It doesn't seem to work very well to estimate 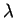 at the same
time as the other hyperparameters,
at the same
time as the other hyperparameters, 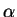 and
and 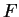 .
POPSPECIFICLAMBDAS estimates a different
.
POPSPECIFICLAMBDAS estimates a different 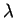 for each
population.
for each
population.
- NOADMIX (Boolean) Assume the model without admixture
(Pritchard et al., 2000a). (Each individual is assumed to be completely
from one of the
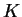 populations.) In the output, instead of printing
the average value of
populations.) In the output, instead of printing
the average value of 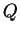 as in the admixture case, the program prints
the posterior probability that each individual is from each
population. 1 = no admixture; 0 = model with admixture.
as in the admixture case, the program prints
the posterior probability that each individual is from each
population. 1 = no admixture; 0 = model with admixture.
- ADMBURNIN (int) (For use when RECOMBINE=1.) When using the
linkage model, a short burnin with the admixture model (say 500
iterations) is strongly recommended in most circumstances. Without
such a burnin, the linkage model often produces peculiar results.
Set
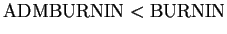 . We have dropped a related
parameter (NOADMBURNIN) that was in Version 1.
. We have dropped a related
parameter (NOADMBURNIN) that was in Version 1.
- USEPOPINFO (Boolean) Use prior population information to
assist clustering. See also MIGRPRIOR and GENSBACK. Must have
POPDATA=1.
- GENSBACK (int) This corresponds to
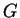 (Pritchard et al., 2000a). When using prior population information for
individuals (USEPOPINFO=1), the program tests whether each individual
has an immigrant ancestor in the last
(Pritchard et al., 2000a). When using prior population information for
individuals (USEPOPINFO=1), the program tests whether each individual
has an immigrant ancestor in the last 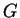 generations, where
generations, where 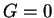 corresponds to the individual being an immigrant itself. In order to
have decent power,
corresponds to the individual being an immigrant itself. In order to
have decent power, 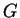 should be set fairly small (2, say) unless the
data are highly informative.
should be set fairly small (2, say) unless the
data are highly informative.
- MIGRPRIOR (double) Must be in [0,1]. This is
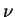 in
Pritchard et al. (2000a). Sensible values might be in the range
0.001--0.1.
in
Pritchard et al. (2000a). Sensible values might be in the range
0.001--0.1.
- PFROMPOPFLAGONLY (Boolean) This option, new with version
2.0, makes it possible to update the allele frequencies,
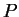 , using
only a prespecified subset of the individuals. To use this, include
a POPFLAG column, and set POPFLAG=1 for individuals who should be
used to update
, using
only a prespecified subset of the individuals. To use this, include
a POPFLAG column, and set POPFLAG=1 for individuals who should be
used to update 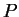 , and POPFLAG=0 for individuals who should not be
used to update
, and POPFLAG=0 for individuals who should not be
used to update 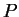 . This can be used both with, or without
USEPOPINFO turned on.
This option will be useful, for example, if you have a standard
reference set of individuals from known populations, and then you
want to estimate the ancestry of some unknown individuals. Using
this option, the
. This can be used both with, or without
USEPOPINFO turned on.
This option will be useful, for example, if you have a standard
reference set of individuals from known populations, and then you
want to estimate the ancestry of some unknown individuals. Using
this option, the 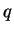 estimate for each unknown individual depends
only on the reference set, and not on the other unknown individuals
in the sample. This property is sometimes desirable.
estimate for each unknown individual depends
only on the reference set, and not on the other unknown individuals
in the sample. This property is sometimes desirable.



Next: Output options
Up: Running structure from the
Previous: Parameters in file mainparams.
Contents
Jonathan Pritchard
2003-07-10