Assignment 1
Medical Images
Overview
In this assignment, you will explore a) classification, b) segmentation, and c) 3d segmentation and semi-supervision on medical images. In part a of the assignment, you will predict severity of diabetic retinopathy in patients using retina images. In part b of the assignment, you will explore segmenting lungs in computerized tomography (CT) scans. In part c of the assignment, you will modify your lung segmentation model to achieve semi-supervised 3D lung segmentation.
Assignment Setup Instructions
-
The starter files will be in the form of Jupyter Notebooks for you to interactively write and run your code. Follow these steps to enable Jupyter Notebook to run on your VM instance:
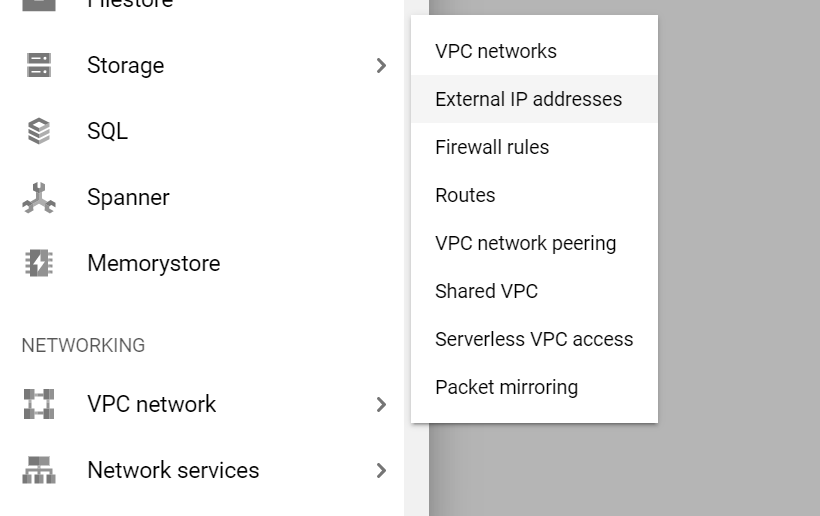
- Navigate to VPC Network > External IP addresses:
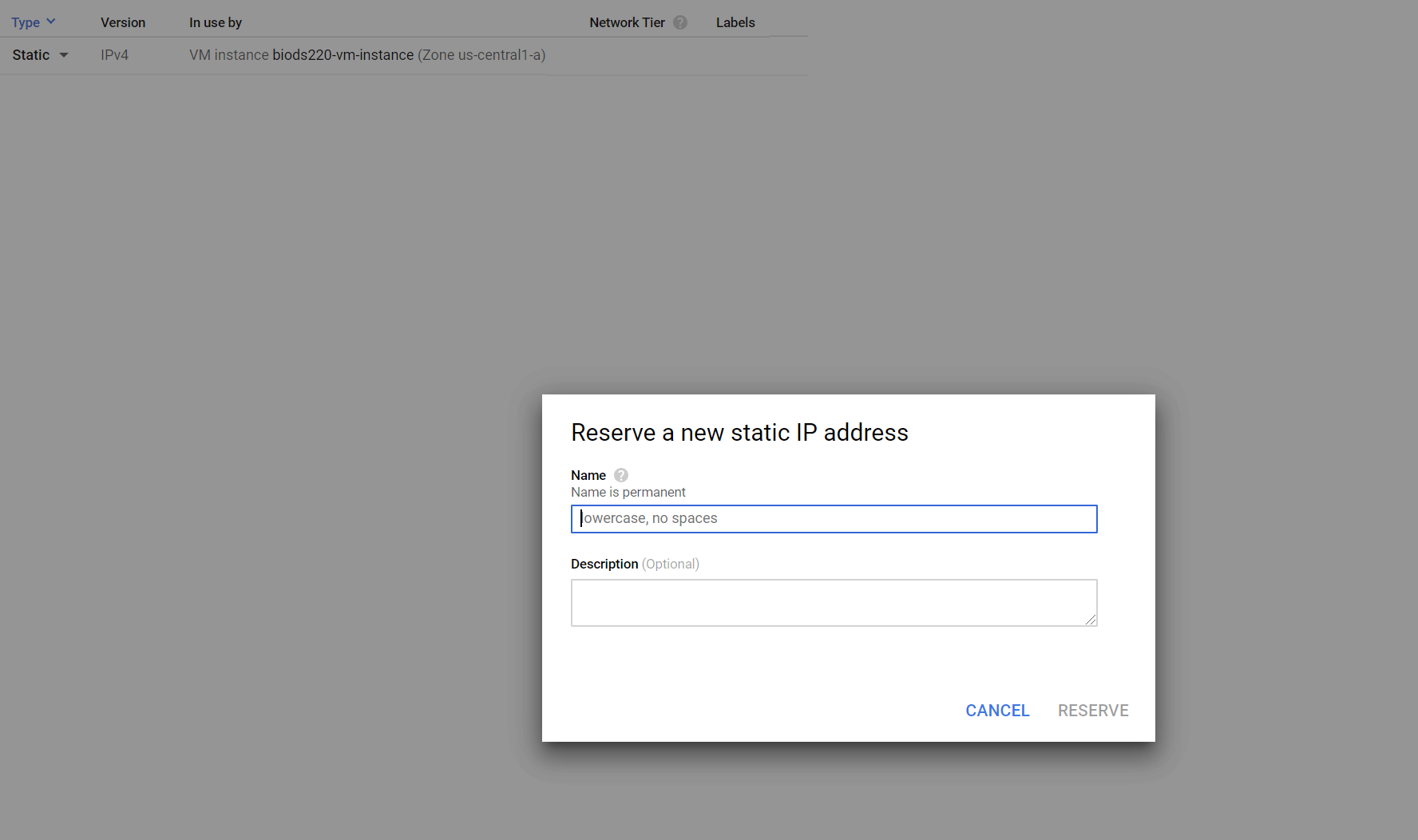
- Make the external IP address static by changing the type, and name it anything you want (e.g. "biods220-ip"). (Note: once you are done using your VM instance for this class, make sure to relinquish the static IP as to not get charged a small fee over time. We will also send out a reminder at the conclusion of the class.)
- Create a Jupyter Notebook configuration file on your VM instance using the following command:
- Edit the config file
~/.jupyter/jupyter_notebook_config.pyand replace the port number with the one you allowed through firewall access (e.g. port 7000) and replace the IP with wildcard (make sure to uncomment both lines as well): - Launch Jupyter Notebook on your VM instance using the following command:
- You can now access the notebook at
http://<external-static-ip-address>:<port-number>on your local machine to complete the assignment and all following assignments. Closing out of the terminal with the open SSH connection will terminate the running Jupyter Notebook.
jupyter notebook --generate-config
(Note: you can use Vim to edit a file on your VM instance. Check these instructions for editing a file in Vim.)c.NotebookApp.ip = '*' c.NotebookApp.port = 7000jupyter notebook
- We will need to increase GCP disk space to fit all the data for this assignment. To do so, please do the following:
- Go to Compute Engine > Disks.
- Click into your VM instance and click EDIT.
- In the Size field, type in "500" GB.
- Click SAVE at the bottom of the page.
- Download the assignment files on your VM instance and then unzip them:
- Now you are all set. Once you unzip, you will notice that there are three notebooks located in
assign1/. Each notebook stands for one part of the assignment - part a) diabetic retinopathy, b) 2D lung segmentation, and c) 3D lung segmentation. Complete both the coding and written sections. To do this, you will need to launch Jupter Notebook as stated above. Good luck on Assignment 1!
wget https://biods220.stanford.edu/downloads/assign1.zip
unzip assign1.zip
Note: In each of Parts 2 and 3 of this assignment, you will be asked to edit your VM instance to switch to using better performing GPUs. To do so, navigate to Compute Engine > VM instances, make sure your VM instance is shut off and click into it, click EDIT, click on CPU platform and GPU and change GPU type based on our instructions.
What to hand in for this assignment: Submit one PDF file containing all your notebook solutions (code + written). To convert your notebook to PDF, go to File > Download as > PDF via LaTeX. Alternatively, if you encounter issues with converting to PDF using LaTeX, you can also convert to HTML (File > Download as > HTML) and then "Save to PDF" by printing. Concatenate the PDFs from all three parts into one and make sure that both the coding sections and the written sections are visible in the PDFs prior to submission. Please make sure when submitting to Gradescope that all questions have been marked with the appropriate pages.