Dominant Drug Targets

Darwinian theory postulates two requirements for evolution to occur. First, populations of individuals must display pre-existing genetic diversity. Then, there must be sufficient selection on survival or reproduction that some members of the population and their progeny become increasingly represented. Viruses often evolve quickly due to the large population sizes, high mutation rates and rapidly changing environments. Dreaded evolutionary events such as adaptation to new hosts or outgrowths of drug-resistant viruses continue to limit human ability to control viral disease.
If a freshly minted genome contains a mutation that confers drug resistance, will it always be selected in the presence of the drug? Not necessarily. During viral infections, newly synthesized viral genomes occupy the same cell as their parent genomes and previously synthesized progeny. If the antiviral target is chosen so that the drug-resistant progeny's growth is dominantly inhibited by the drug-susceptible members of its intracellular family, the outgrowth of otherwise fit drug-resistant genomes can be suppressed. Precedent for 'dominant drug targeting' as a deliberate approach to suppress the outgrowth of inhibitor-resistant viruses has been established for capsid variants of foot-and-mouth disease virus, poliovirus (see image) and dengue virus. Our laboratory is focused on identification of additional dominant drug targets for antiviral design. Small molecules that stabilize oligomeric assemblages are a promising means to confer to any unfit family the ability to destroy the effectiveness of a newborn relative with increased fitness via co-assembly of drug-susceptible and drug-resistant monomers. Most positive strand RNA viruses could be inhibited by dominant drug targeting including HCV, Zika virus, DNV and Influenza viruses.
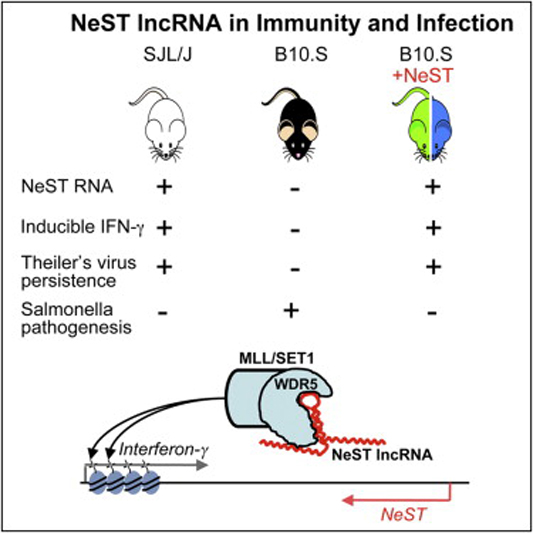
NeST lncRNA
cDNA sequencing analysis suggests that at least 10,000 long non-coding RNAs (lncRNAs) are produced from the intergenic regions of the human chromosome. Only a handful of these lncRNAs have been characterized to date; the NeST RNA was the first to be linked with an immune phenotype. NeST was discovered as the genetic basis of a susceptibility locus originally mapped by its effects of the multigenic control to viral and bacterial infections. It is highly polymorphic in laboratory inbred mouse strains suggesting it has been under purifying selection by challenges to the immune response. In humans, the NeST-encoding regions are the site of disease-associated alleles for various immunological diseases that include Rheumatoid arthritis, asthma and inflammatory bowl disease. NeST expression has been observed in CD4+ Th1 cells, CD8+ T cells, and NK cells and is protective to systemic bacterial infections. It functions in the induction of IFNγ in T cells through its effects on chromatin methylation. The mechanisms that lead to the control of inflammatory microbial challenges by NeST RNA are a major focus in our laboratory.
Previous attempts to understand epigenetic functions of lncRNAs in the immune response have relied on techniques that require large numbers of cells, and as a result, these studies have solely been performed in the context of cultured cells in vitro. Therefore, the importance of these molecules in the context of a systems-level immune response in vivo remains somewhat uncertain. Our laboratory is currently leveraging our expertise in immunology, epigenome analysis, NeST biology and host-pathogen interactions to understand the function of lncRNAs in the immune response in vivo.
Autophagy and Virus Infections

Autophagy is a mechanism by which eukaryotic cells degrade cytosolic components, targeting them for degradation via the formation of unique double-membraned vesicles that then enter the endosomal/lysosomal pathway. Autophagy is recently appreciated to be an important component of the innate immune response in both vertebrates and invertebrates, directly destroying many intracellular pathogens. With this in mind, compounds that stimulate autophagy promise to be valuable immunomodulatory therapeutics for a wide variety of infections. However, some pathogens, including several RNA viruses, appear to subvert the autophagy pathway to facilitate their replication and, possibly, intercellular spread. For these pathogens, inhibitors of autophagy could be very valuable therapeutics. For these scenarios to become reality, a basic understanding of how the autophagy pathway can, in some cases, clear infections while promoting others would be highly valuable.