


Next: Running simulations.
Up: Front End
Previous: Building a project.
Contents
Once you've successfully loaded a data file, you are ready to start
running structure. You will create one or more ``parameter
sets''; these represent a whole list of choices that you make about
how to analyze the data. We have entered a series of default settings,
and these are good place to start. You will probably want to run
structure multiple times for each parameter set, at different
values of 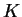 , and the front end is set up to facilitate this.
Go to the pull-down menu under Parameter Set. You can create a
new parameter set, modify an existing one, or delete one. Click on
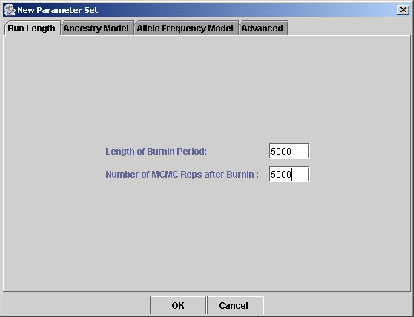
``New''. You now see a dialogue box with four tabs (Figure 5).
Click on each of these:
, and the front end is set up to facilitate this.
Go to the pull-down menu under Parameter Set. You can create a
new parameter set, modify an existing one, or delete one. Click on
``New''. You now see a dialogue box with four tabs (Figure 5).
Click on each of these:
- Run Length. See section 4.1 for discussion of this.
Note that the front end provides time series plots of some key parameters
to help you assess whether the run length seems to be sufficient.
- Ancestry Model. See section AncestryModels. The
admixture model is a good place to start for most data sets. The
linkage model will be disabled unless you entered linkage information
about the markers. Section 7 provides some extra details
about some of the detailed options, including GENSBACK and MIGRPRIOR
under the ``Use Population Information'' option. Note that the linkage
model is relatively computationally intensive.
- Allele Frequency Model. See section
4.3. We recommend applying both the correlated
frequencies model and the independent frequencies model. The
correlated frequencies model has better power to detect subtle
population structure, but the posterior probabilities for
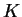 may be
biased upwards somewhat. The correlated frequencies model is
parameterized in terms of
may be
biased upwards somewhat. The correlated frequencies model is
parameterized in terms of 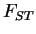 , with a separate parameter for each
population (details in section 4.3). Inferring
, with a separate parameter for each
population (details in section 4.3). Inferring
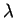 is probably not necessary during initial investigation.
is probably not necessary during initial investigation.
- Advanced. Turning off the function that computes the
posterior probabilities (for estimating
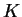 ) speeds the program up
significantly. You can also get the program to output posterior
credible regions for the ancestry of each individual (see ANCESTDIST,
section 7.3). ``Initialize at POPINFO'' is
described in more detail under STARTATPOPINFO, section 7.5.
) speeds the program up
significantly. You can also get the program to output posterior
credible regions for the ancestry of each individual (see ANCESTDIST,
section 7.3). ``Initialize at POPINFO'' is
described in more detail under STARTATPOPINFO, section 7.5.
Figure 5:
Specifying a new parameter set: setting the run length.
 |



Next: Running simulations.
Up: Front End
Previous: Building a project.
Contents
Jonathan Pritchard
2003-07-10