Single-Molecule Projects
Trapping Single Photosynthetic Proteins and Enzymes in Solution: The Anti-Brownian ELectrokinetic
(ABEL) Trap
Subgroup Members: Dr. William (Memo) Carpenter, Abhijit Lavania
Support: Department of Energy; Office of Science, Basic Energy Sciences; Chemical Sciences, Geosciences, and Biosciences Division, Physical Biosciences Area
Basics
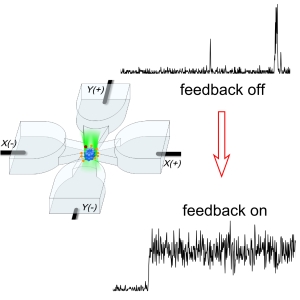 We have developed a new type of trap for nanoscale particles in solution,
the ABEL trap. Here's the key ideas: We have developed a new type of trap for nanoscale particles in solution,
the ABEL trap. Here's the key ideas:
Nanoscale objects in solution (such as proteins, enzymes, and DNA) are
continually bombarded by thermally agitated molecules of solvent.
This bombardment makes nanoscale objects jiggle around in solution. The smaller
an object is, the faster (and farther) it jiggles. This jiggling,
also known as Brownian motion, makes the task of studying nano-objects
in solution very difficult: the objects just don't hold still.
The Anti-Brownian Electrokinetic trap (ABEL trap) eliminates
the Brownian motion of one object in solution, allowing detailed
examination of its properties. The ABEL trap works by carefully measuring
the tiny Brownian displacements of the object, and then using
a feedback loop to apply electric fields to the solution to exactly
cancel these displacements using electrokinetic forces, which may arise from electrophoresis or electroosmosis. This is all done in a microfluidic geometry illustrated in the figure.
To trap very tiny objects, we have to apply the feedback extremely
quickly--if there is too much delay, the object escapes before
the ABEL trap can bring it back to the target position. Using a variety of approaches, the position update and consequent feedback application time has been made fast enough to trap individual fluorescently
labeled protein molecules in solution. When this was first done, these molecules (as small
as 10 nm in diameter) were the first proteins trapped in solution
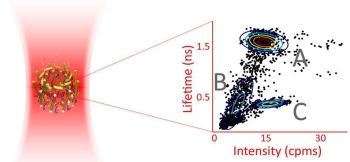
and the smallest objects ever trapped in solution. More importantly, we trap the single biomolecules in a region of time-averaged uniform intensity, so that the brightness of the object becomes a useful measurement variable (see figure lower right panel). Compare with fluorescence correlation spectroscopy (FCS), where infomration is obtained from individual ms-long bursts of fluorescence when a biomolecule diffuses through an ellipsoidal-shaped laser focal spot. This achievement
opens the possibility of studying the detailed dynamical behavior of individual proteins free-floating
in solution where many variables can be measured such as brightness, degree of FRET, lifetime, and emission spectrum.
The ABEL trap gives scientists a new handle on the nano-world.
This idea should have applications in diverse areas, including
the trapping and study of single biomolecules, nanoscale fabrication,
and statistical physics/mechanics. Current projects are addressing conformational changes and enzymatic kinetics in trapped enzymes, photophysics/photodynamics of antenna proteins, measurements of aggregation states of photosynthetic proteins, and the counting of protein subunits.
As a side note, it is interesting to have first demonstrated this trap in 2005, 100 years after Einstein's annus mirabilis,
in which one of his key papers of that year addressed the fundamental thermal mechanisms
for Brownian motion: Annalen der Physik 17, 549 (1905).
Recent reviews:
William Carpenter, Abhijit Lavania, Allison Squires, and W.E. Moerner, “Label-free anti-Brownian trapping of single nanoparticles in solution,” Invited Article, Special Issue of J Phys Chem C 128, 20275-20286 (2024), Part of Photothermal and Non-Fluorescent Imaging in Microscopy and Spectroscopy VSI Virtual Special Issue, (DOI: 10.1021/acs.jpcc.4c05878 published online 19 November 2024)
Alison H. Squires, Quan Wang, Peter D. Dahlberg, and W. E. Moerner, “A bottom-up perspective on photodynamics and photoprotection in light-harvesting complexes using anti-Brownian trapping,” J. Chem. Phys. 156, 070901 (2022) (DOI: 10.1063/5.0079042, published online 16 February 2022).
Allison H. Squires, Adam E. Cohen, and W. E. Moerner, “Anti-Brownian Traps,” in G. C. K. Roberts, A. Watts, European Biophysical Societies (eds.), Encyclopedia of Biophysics. Springer, Berlin, Heidelberg, 2018. (DOI: 10.1007/978-3-642-35943-9_486-1)
Gabriela S. Schlau-Cohen, Samuel Bockenhauer, Quan Wang, and W. E. Moerner, “Single-molecule spectroscopy of photosynthetic proteins in solution: exploration of structure–function relationships,” Minireview, Chem. Sci. 5, 2933-2939 (DOI:10.1039/C4SC00582A, published online 15 April 2014). DOI
Quan Wang, Randall H. Goldsmith, Yan Jiang, Samuel D. Bockenhauer, and W.E. Moerner, “Probing single biomolecules in solution using the Anti-Brownian ELectrokinetic (ABEL) trap,” Acc. Chem. Res. (Paul Barbara Special Issue) 45, 1955-1964 (2012), published online 22 May 2012. DOI
A. E. Cohen and W. E. Moerner, “Anti-Brownian Traps,” in Encyclopedia of Biophysics, G. C. K. Roberts (Ed.) (Springer, Berlin, Heidelberg, 2012), pp. 95-97.
Recent Advances:
Exploring Masses and Internal Mass Distributions of Single Carboxysomes in Free Solution Using Fluorescence and Interferometric Scattering in an Anti-Brownian Trap
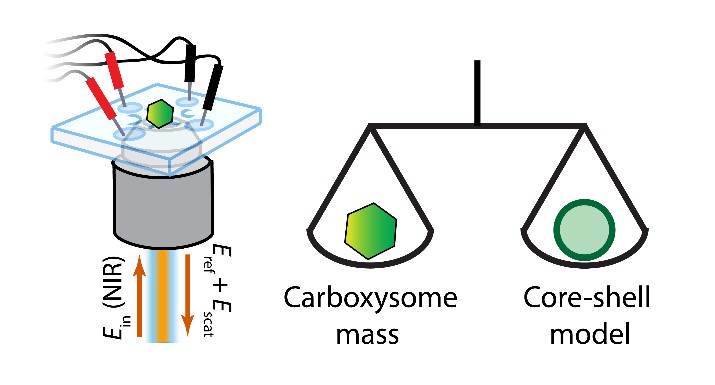 Carboxysomes are self-assembled bacterial microcompartments that facilitate carbon assimilation by co-localizing the enzymes of CO2 fixation within a protein shell. These microcompartments can be highly heterogeneous in their composition and filling, so measuring the mass and loading of an individual carboxysome would allow for better characterization of its assembly and function. To enable detailed and extended characterizations of single nanoparticles in solution, we recently demonstrated an improved Interferometric Scattering Anti-Brownian ELectrokinetic (ISABEL) trap, which tracks the position of a single nanoparticle via its scattering of a near-infrared beam and applies feedback to counteract its Brownian motion. Importantly, the scattering signal can be related to the mass of nanoscale proteinaceous objects, whose refractive indices are well-characterized. We calibrate single-particle scattering cross-section measurements in the ISABEL trap and determine individual carboxysome masses in the 50-400 MDa range by analyzing their scattering cross-sections with a core-shell model. We further investigate carboxysome loading by combining mass measurements with simultaneous fluorescence reporting from labeled internal components. This method may be extended to other biological objects, such as viruses or extracellular vesicles, and can be combined with orthogonal fluorescence reporters to achieve precise physical and chemical characterization of individual nanoscale biological objects. Carboxysomes are self-assembled bacterial microcompartments that facilitate carbon assimilation by co-localizing the enzymes of CO2 fixation within a protein shell. These microcompartments can be highly heterogeneous in their composition and filling, so measuring the mass and loading of an individual carboxysome would allow for better characterization of its assembly and function. To enable detailed and extended characterizations of single nanoparticles in solution, we recently demonstrated an improved Interferometric Scattering Anti-Brownian ELectrokinetic (ISABEL) trap, which tracks the position of a single nanoparticle via its scattering of a near-infrared beam and applies feedback to counteract its Brownian motion. Importantly, the scattering signal can be related to the mass of nanoscale proteinaceous objects, whose refractive indices are well-characterized. We calibrate single-particle scattering cross-section measurements in the ISABEL trap and determine individual carboxysome masses in the 50-400 MDa range by analyzing their scattering cross-sections with a core-shell model. We further investigate carboxysome loading by combining mass measurements with simultaneous fluorescence reporting from labeled internal components. This method may be extended to other biological objects, such as viruses or extracellular vesicles, and can be combined with orthogonal fluorescence reporters to achieve precise physical and chemical characterization of individual nanoscale biological objects.
Abhijit A. Lavania, William B. Carpenter, Luke M. Oltrogge, Davis Perez, Julia B. Turns?ek, David F. Savage, and W. E. Moerner, “Exploring Masses and Internal Mass Distributions of Single Carboxysomes in Free Solution Using Fluorescence and Interferometric Scattering in an Anti-Brownian Trap,” Part of Special Issue “Steven G. Boxer Festschrift,” J. Phys. Chem. B 126, 8747-8759 (2022) (DOI: 10.1021/acs.jpcb.2c05939, published online 25 October 2022).
Ratiometric Sensing of Redox Environments Inside Individual Carboxysomes Trapped in Solution
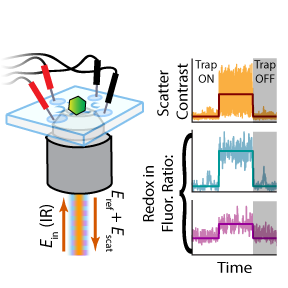 Diffusion of biological nanoparticles in solution impedes our ability to continuously monitor individuals and measure their physical and chemical properties. To overcome this, we previously developed the Interferometric Scattering Anti-Brownian ELectrokinetic (ISABEL) trap, which uses scattering to localize a particle and applies electrokinetic forces which counteract Brownian motion, thus enabling extended observation. Here, we present an improved ISABEL trap that incorporates a near-infrared scatter illumination beam and rapidly interleaves 405 and 488 nm fluorescence excitation reporter beams. With the ISABEL trap, we monitor the internal redox environment of individual carboxysomes labeled with the ratiometric redox reporter roGFP2. Carboxysomes widely vary in scattering contrast (reporting on size) and redox-dependent ratiometric fluorescence. Further, we used redox sensing to explore the chemical kinetics within intact carboxysomes, where bulk measurements may contain unwanted contributions from aggregates or interfering fluorescent proteins. Overall, we demonstrate the ISABEL trap’s ability to sensitively monitor nanoscale biological objects, enabling new experiments on these systems. Diffusion of biological nanoparticles in solution impedes our ability to continuously monitor individuals and measure their physical and chemical properties. To overcome this, we previously developed the Interferometric Scattering Anti-Brownian ELectrokinetic (ISABEL) trap, which uses scattering to localize a particle and applies electrokinetic forces which counteract Brownian motion, thus enabling extended observation. Here, we present an improved ISABEL trap that incorporates a near-infrared scatter illumination beam and rapidly interleaves 405 and 488 nm fluorescence excitation reporter beams. With the ISABEL trap, we monitor the internal redox environment of individual carboxysomes labeled with the ratiometric redox reporter roGFP2. Carboxysomes widely vary in scattering contrast (reporting on size) and redox-dependent ratiometric fluorescence. Further, we used redox sensing to explore the chemical kinetics within intact carboxysomes, where bulk measurements may contain unwanted contributions from aggregates or interfering fluorescent proteins. Overall, we demonstrate the ISABEL trap’s ability to sensitively monitor nanoscale biological objects, enabling new experiments on these systems.
William B. Carpenter, Abhijit A. Lavania, Julia S. Borden, Luke M. Oltrogge, Davis Perez, Peter D. Dahlberg, David F. Savage, and W. E. Moerner, “Ratiometric Sensing of Redox Environments Inside Individual Carboxysomes Trapped in Solution," J. Phys. Chem Lett. 13, 4455-4462 (2022) (DOI: 10.1021/acs.jpclett.2c00782, published online May 13, 2022).
Interferometric scattering enables fluorescence-free electrokinetic trapping of single nanoparticles in free solution
 Anti-Brownian traps confine single particles in free solution by closed-loop feedback forces that directly counteract Brownian motion. The extended-duration measurement of trapped objects allows detailed characterization of photophysical and transport properties, as well as observation of infrequent or rare dynamics. However, this approach has been generally limited to particles that can be tracked by fluorescent emission. Here we present the Interferometric Scattering Anti-Brownian ELectrokinetic trap (ISABEL trap), which uses interferometric scattering rather than fluorescence to monitor particle position. By decoupling the ability to track (and therefore trap) a particle from collection of its spectroscopic data, the ISABEL trap enables confinement and extended study of single particles that do not fluoresce, that only weakly fluoresce, or which exhibit intermittent fluorescence or photobleaching. This new technique significantly expands the range of nanoscale objects that may be investigated at the single-particle level in free solution. Anti-Brownian traps confine single particles in free solution by closed-loop feedback forces that directly counteract Brownian motion. The extended-duration measurement of trapped objects allows detailed characterization of photophysical and transport properties, as well as observation of infrequent or rare dynamics. However, this approach has been generally limited to particles that can be tracked by fluorescent emission. Here we present the Interferometric Scattering Anti-Brownian ELectrokinetic trap (ISABEL trap), which uses interferometric scattering rather than fluorescence to monitor particle position. By decoupling the ability to track (and therefore trap) a particle from collection of its spectroscopic data, the ISABEL trap enables confinement and extended study of single particles that do not fluoresce, that only weakly fluoresce, or which exhibit intermittent fluorescence or photobleaching. This new technique significantly expands the range of nanoscale objects that may be investigated at the single-particle level in free solution.
Allison H. Squires, Abhijit A. Lavania, Peter D. Dahlberg, and W.E. Moerner, “Interferometric scattering enables fluorescence-free electrokinetic trapping of single nanoparticles in free solution,” Nano Letters 19, 4112-4117 (2019) (DOI: 10.1021/acs.nanolett.9b01514, published online 22 May 2019). DOI [ Slide] Slide]
Also see Abhijit A. Lavania, Allison H. Squires, Peter D. Dahlberg, W. E. Moerner, "Interferometric scattering for fluorescence-free electrokinetic trapping of single nanoparticles in free solution," Proc. SPIE 11246, Single Molecule Spectroscopy and Superresolution Imaging XIII, 112460W (13 February 2020) (DOI: 10.1117/12.2546638).
Single-molecule trapping and spectroscopy reveals photophysical heterogeneity of phycobilisomes quenched by Orange Carotenoid Protein
 The Orange Carotenoid Protein (OCP) is a cytosolic photosensor that is responsible for non-photochemical quenching (NPQ) of the light-harvesting process in most cyanobacteria. Uponphotoactivation by blue-green light, OCP binds to the phycobilisome antenna complex,providing an excitonic trap to thermally dissipate excess energy. At present, both the bindingsite and NPQ mechanism of OCP are unknown. Using an Anti-Brownian ELectrokinetic(ABEL) trap, we isolate single phycobilisomes in free solution, both in the presence andabsence of activated OCP, to directly determine the photophysics and heterogeneity of OCP-quenched phycobilisomes. Surprisingly, we observe two distinct OCP-quenched states, withlifetimes 0.09 ns (6% of unquenched brightness) and 0.21 ns (11% brightness). Photon-by-photon Monte Carlo simulations of exciton transfer through the phycobilisome suggest thatthe observed quenched states are kinetically consistent with either two or one bound OCPs,respectively, underscoring an additional mechanism for excitation control in this key pho-tosynthetic unit The Orange Carotenoid Protein (OCP) is a cytosolic photosensor that is responsible for non-photochemical quenching (NPQ) of the light-harvesting process in most cyanobacteria. Uponphotoactivation by blue-green light, OCP binds to the phycobilisome antenna complex,providing an excitonic trap to thermally dissipate excess energy. At present, both the bindingsite and NPQ mechanism of OCP are unknown. Using an Anti-Brownian ELectrokinetic(ABEL) trap, we isolate single phycobilisomes in free solution, both in the presence andabsence of activated OCP, to directly determine the photophysics and heterogeneity of OCP-quenched phycobilisomes. Surprisingly, we observe two distinct OCP-quenched states, withlifetimes 0.09 ns (6% of unquenched brightness) and 0.21 ns (11% brightness). Photon-by-photon Monte Carlo simulations of exciton transfer through the phycobilisome suggest thatthe observed quenched states are kinetically consistent with either two or one bound OCPs,respectively, underscoring an additional mechanism for excitation control in this key pho-tosynthetic unit
Allison H. Squires, Peter D. Dahlberg, Haijun Liu, Nikki Cecil M. Magdaong, Robert E. Blankenship, and W. E. Moerner, “Single-molecule trapping and spectroscopy reveals photophysical heterogeneity of phycobilisomes quenched by Orange Carotenoid Protein.” Nature Commun. 10, article 1172 (2019) (DOI: 10.1038/s41467-019-09084-2, published online 12 March 2019). DOI [ Slide] Slide]
Resolving Mixtures in Solution by Single-Molecule Rotational Diffusivity
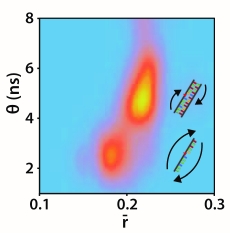 Sensing the size of individual molecules in an ensemble has proven to be a powerful tool to investigate biomolecular interactions and association-dissociation processes. In biologically-relevant solution environments, molecular size is often sensed by translational or rotational diffusivity. The rotational diffusivity is more sensitive to the size and conformation of the molecules as it is inversely proportional to the cube of the hydrodynamic radius, as opposed to the inverse linear dependence of the translational diffusion coefficient. Single-molecule rotational diffusivity has been measured with time-resolved fluorescence anisotropy decay, but the ability to sense different sizes has been restricted by the limited number of photons available or has required surface attachment to observe each molecule longer, and the attachment may be perturbative. To address these limitations, we show how to measure and monitor single-molecule rotational diffusivity by combining the solution-phase Anti-Brownian ELectrokinetic (ABEL) trap and maximum likelihood analysis of time-resolved fluorescence anisotropy based on the information inherent in each detected photon. We demonstrate this approach by resolving a mixture of single- and double-stranded fluorescently-labeled DNA molecules at equilibrium, freely rotating in a native solution environment. The rotational diffusivity, fluorescence brightness and lifetime, and initial and steady-state anisotropy, are simultaneously determined for each trapped single DNA molecule. The time resolution and precision of this method were analyzed using statistical signal analysis and simulations. We present key parameters that define the usefulness of a particular fluorescent label for extracting molecular size information from single-molecule rotational diffusivity. Sensing the size of individual molecules in an ensemble has proven to be a powerful tool to investigate biomolecular interactions and association-dissociation processes. In biologically-relevant solution environments, molecular size is often sensed by translational or rotational diffusivity. The rotational diffusivity is more sensitive to the size and conformation of the molecules as it is inversely proportional to the cube of the hydrodynamic radius, as opposed to the inverse linear dependence of the translational diffusion coefficient. Single-molecule rotational diffusivity has been measured with time-resolved fluorescence anisotropy decay, but the ability to sense different sizes has been restricted by the limited number of photons available or has required surface attachment to observe each molecule longer, and the attachment may be perturbative. To address these limitations, we show how to measure and monitor single-molecule rotational diffusivity by combining the solution-phase Anti-Brownian ELectrokinetic (ABEL) trap and maximum likelihood analysis of time-resolved fluorescence anisotropy based on the information inherent in each detected photon. We demonstrate this approach by resolving a mixture of single- and double-stranded fluorescently-labeled DNA molecules at equilibrium, freely rotating in a native solution environment. The rotational diffusivity, fluorescence brightness and lifetime, and initial and steady-state anisotropy, are simultaneously determined for each trapped single DNA molecule. The time resolution and precision of this method were analyzed using statistical signal analysis and simulations. We present key parameters that define the usefulness of a particular fluorescent label for extracting molecular size information from single-molecule rotational diffusivity.
Hsiang-Yu Yang and W. E. Moerner, “Resolving Mixtures in Solution by Single-Molecule Rotational Diffusivity,” Nano Lett. 18, 5279-5287 (2018) (DOI: 10.1021/acs.nanolett.8b02280, published online 12 July 2018). DOI
Direct single-molecule measurements of phycocyanobilin photophysics for the monomeric C-phycocyanin pigment-protein antenna complex
 Phycobilisomes are highly organized pigment–protein antenna complexes found in the photosynthetic apparatus of cyanobacteria and rhodophyta that harvest solar energy and transport it to the reaction center. A detailed bottom-up model of pigment organization and energy transfer in phycobilisomes is essential to understanding photosynthesis in these organisms and informing rational design of artificial light-harvesting systems. In particular, heterogeneous photophysical behaviors of these proteins, which cannot be predicted de novo, may play an essential role in rapid light adaptation and photoprotection. Furthermore, the delicate architecture of these pigment–protein scaffolds sensitizes them to external perturbations, for example, surface attachment, which can be avoided by study in free solution or in vivo. Here, we present single-molecule characterization of C-phycocyanin (C-PC), a threepigment biliprotein that self-assembles to form the midantenna rods of cyanobacterial phycobilisomes. Using the Anti-Brownian Electrokinetic (ABEL) trap to counteract Brownian motion of single particles in real time, we directly monitor the changing photophysical states of individual C-PC monomers from Spirulina platensis in free solution by simultaneous readout of their brightness, fluorescence anisotropy, fluorescence lifetime, and emission spectra. These include single-chromophore emission states for each of the three covalently bound phycocyanobilins, providing direct measurements of the spectra and photophysics of these chemically identical molecules in their native protein environment. We further show that a simple Förster resonant energy transfer (FRET) network model accurately predicts the observed photophysical states of C-PC and suggests highly variable quenching behavior of one of the chromophores, which should inform future studies of higherorder complexes. Phycobilisomes are highly organized pigment–protein antenna complexes found in the photosynthetic apparatus of cyanobacteria and rhodophyta that harvest solar energy and transport it to the reaction center. A detailed bottom-up model of pigment organization and energy transfer in phycobilisomes is essential to understanding photosynthesis in these organisms and informing rational design of artificial light-harvesting systems. In particular, heterogeneous photophysical behaviors of these proteins, which cannot be predicted de novo, may play an essential role in rapid light adaptation and photoprotection. Furthermore, the delicate architecture of these pigment–protein scaffolds sensitizes them to external perturbations, for example, surface attachment, which can be avoided by study in free solution or in vivo. Here, we present single-molecule characterization of C-phycocyanin (C-PC), a threepigment biliprotein that self-assembles to form the midantenna rods of cyanobacterial phycobilisomes. Using the Anti-Brownian Electrokinetic (ABEL) trap to counteract Brownian motion of single particles in real time, we directly monitor the changing photophysical states of individual C-PC monomers from Spirulina platensis in free solution by simultaneous readout of their brightness, fluorescence anisotropy, fluorescence lifetime, and emission spectra. These include single-chromophore emission states for each of the three covalently bound phycocyanobilins, providing direct measurements of the spectra and photophysics of these chemically identical molecules in their native protein environment. We further show that a simple Förster resonant energy transfer (FRET) network model accurately predicts the observed photophysical states of C-PC and suggests highly variable quenching behavior of one of the chromophores, which should inform future studies of higherorder complexes.
Allison H. Squires and W. E. Moerner, “Direct single-molecule measurements of phycocyanobilim photophysics for monomeric C-phycocyanin," Proc. Nat. Acad. Sci. (USA) (2017) (DOI: 10.1073/pnas.1705435114, published online 28 August 2017). DOI [ Slide] Slide]
Exploring the Pigment Architecture and Photodynamics of the Allophycocyanin Monomer and Trimer Using Multivariate Measurements of Single Molecules in Solution
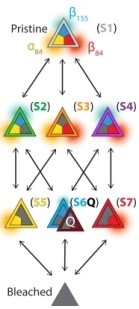
 Oligomerization plays a critical role in shaping the light-harvesting properties of many photosynthetic pigment-protein complexes, but a detailed understanding of this process at the level of individual pigments is still lacking. To study the effects of oligomerization, we designed a single-molecule approach to probe the photophysical properties of individual pigment sites as a function of protein assembly state. Our method, based on the principles of anti-Brownian electrokinetic trapping of single fluorescent proteins, step-wise photobleaching, and multi-parameter spectroscopy, allows pigment-specific spectroscopic information on single multi-pigment antennae to be recorded in a non-perturbative aqueous environment with unprecedented detail. We focus on the monomer-to-trimer transformation of allophycocyanin (APC), an important antenna protein in cyanobacteria. Our data reveals that the two chemically-identical pigments in APC have different roles. One (α) is the functional pigment that red-shifts its spectral properties upon trimer formation, while the other (β) is a “protective” pigment that persistently quenches the excited state of α in the pre-functional, monomer state of the protein. These results show how subtleties in pigment organization give rise to functionally important aspects of energy transfer and photoprotection in antenna complexes. The method developed here should find immediate application in understanding the emergent properties of other natural and artificial light-harvesting systems. Oligomerization plays a critical role in shaping the light-harvesting properties of many photosynthetic pigment-protein complexes, but a detailed understanding of this process at the level of individual pigments is still lacking. To study the effects of oligomerization, we designed a single-molecule approach to probe the photophysical properties of individual pigment sites as a function of protein assembly state. Our method, based on the principles of anti-Brownian electrokinetic trapping of single fluorescent proteins, step-wise photobleaching, and multi-parameter spectroscopy, allows pigment-specific spectroscopic information on single multi-pigment antennae to be recorded in a non-perturbative aqueous environment with unprecedented detail. We focus on the monomer-to-trimer transformation of allophycocyanin (APC), an important antenna protein in cyanobacteria. Our data reveals that the two chemically-identical pigments in APC have different roles. One (α) is the functional pigment that red-shifts its spectral properties upon trimer formation, while the other (β) is a “protective” pigment that persistently quenches the excited state of α in the pre-functional, monomer state of the protein. These results show how subtleties in pigment organization give rise to functionally important aspects of energy transfer and photoprotection in antenna complexes. The method developed here should find immediate application in understanding the emergent properties of other natural and artificial light-harvesting systems.
See Highlight by Robert Blankenship.
Quan Wang and W. E. Moerner, “Dissecting pigment architecture of individual photosynthetic antenna complexes in solution,” Proc. Nat. Acad. Sci. (USA) 112, 13880-13885 (2015) (DOI: 10.1073/pnas.1514027112, published online October 5, 2015). DOI [ Slide] Slide]
Single-Molecule Identification of Quenched and Unquenched States of the primary light-harvesting complex in green plants, LHCII
 In photosynthetic light harvesting, absorbed sunlight is converted to electron flow with near-unity quantum efficiency. Under high light conditions, plants avoid damage to their molecular machinery by activating a set of photoprotective mechanisms to harmlessly dissipate excess energy as heat. To investigate these mechanisms, we study the primary antenna complex in green plants, light-harvesting complex II (LHCII), at the single-complex level. We use a single-molecule technique, the Anti-Brownian Electrokinetic (ABEL) trap, which enables simultaneous measurements of fluorescence intensity, lifetime, and spectra in solution. With this approach, including the first measurements of fluorescence lifetime on single LHCII complexes, we access the intrinsic conformational dynamics. In addition to an unquenched state, we identify two partially quenched states of LHCII. Our results suggest that there are at least two distinct quenching sites with different molecular compositions, meaning multiple dissipative pathways in LHCII. Furthermore, one of the quenched conformations significantly increasesin relative population under environmental conditions mimicking high light. In photosynthetic light harvesting, absorbed sunlight is converted to electron flow with near-unity quantum efficiency. Under high light conditions, plants avoid damage to their molecular machinery by activating a set of photoprotective mechanisms to harmlessly dissipate excess energy as heat. To investigate these mechanisms, we study the primary antenna complex in green plants, light-harvesting complex II (LHCII), at the single-complex level. We use a single-molecule technique, the Anti-Brownian Electrokinetic (ABEL) trap, which enables simultaneous measurements of fluorescence intensity, lifetime, and spectra in solution. With this approach, including the first measurements of fluorescence lifetime on single LHCII complexes, we access the intrinsic conformational dynamics. In addition to an unquenched state, we identify two partially quenched states of LHCII. Our results suggest that there are at least two distinct quenching sites with different molecular compositions, meaning multiple dissipative pathways in LHCII. Furthermore, one of the quenched conformations significantly increasesin relative population under environmental conditions mimicking high light.
Gabriela S. Schlau-Cohen, Hsiang-Yu Yang, Tjaart P. J. Krüger, Pengqi Xu, Michal Gwizdala, Rienk van Grondelle, Roberta Croce, and W. E. Moerner, “Single-Molecule Identification of Quenched and Unquenched States of LHCII,” J. Phys. Chem. Lett. 6, 860-867 (2015). (DOI: 10.1021/acs.jpclett.5b00034, publ. online February 18, 2015). DOI [ Slide] Slide]
Single-molecule motions enable direct visualization of biomolecular interactions in solution
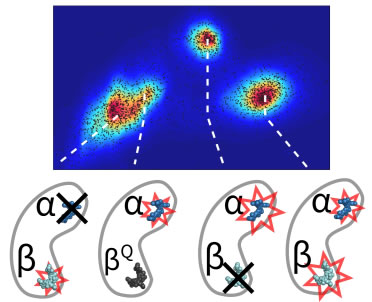
 Most single-molecule studies infer the behavior of the system from changes in the emission properties of a fluorescent label, such as brightness, excited state lifetime, or spectrum. At the same time, biomolecular interactions are generally accompanied by modifications in the size and charge of biomolecules at the nanometer scale, but these do not in general lead to changes in the emission of a fluorescent label. Here we describe a new and advanced ABEL trap method to sense these changes in real time based on statistical learning of diffusive and electric field–induced motion parameters of a trapped molecule in solution. Essentially, the analysis system measures the residual motion of the trapped single molecule, which contains information about its diffusion and its response to the trapping fields. In this way, new single-molecule variables are made available, which provide new ways to distinguish different molecules in solution, as well as to observe their dynamical changes. We demonstrate the approach by resolving a monomer-trimer mixture along a protein dissociation pathway and by visualizing the binding-unbinding kinetics of a single DNA molecule. Most single-molecule studies infer the behavior of the system from changes in the emission properties of a fluorescent label, such as brightness, excited state lifetime, or spectrum. At the same time, biomolecular interactions are generally accompanied by modifications in the size and charge of biomolecules at the nanometer scale, but these do not in general lead to changes in the emission of a fluorescent label. Here we describe a new and advanced ABEL trap method to sense these changes in real time based on statistical learning of diffusive and electric field–induced motion parameters of a trapped molecule in solution. Essentially, the analysis system measures the residual motion of the trapped single molecule, which contains information about its diffusion and its response to the trapping fields. In this way, new single-molecule variables are made available, which provide new ways to distinguish different molecules in solution, as well as to observe their dynamical changes. We demonstrate the approach by resolving a monomer-trimer mixture along a protein dissociation pathway and by visualizing the binding-unbinding kinetics of a single DNA molecule.
Quan Wang and W.E. Moerner, "Single-molecule motions enable direct visualization of biomolecular interactions in solution," Nature Methods111, 555-558 (2014), published online 9 March 2014. DOI [ Slide] Slide]
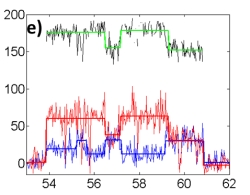
Single-molecule spectroscopy in the ABEL trap reveals LH2 complexes switch between emissive states
 Photosynthetic organisms flourish under low light intensities by converting photoenergy to chemical energy with near unity quantum efficiency and under high light intensities by safely dissipating excess photoenergy and deleterious photoproducts. The molecular mechanisms balancing these two functions remains incompletely described. One critical barrier to characterizing the mechanisms responsible for these processes is that they occur within proteins whose excited-state properties vary drastically between individual proteins and even within a single protein over time. In ensemble measurements, these excited-state properties appear only as the average value. To overcome this averaging, we investigate the purple bacterial antenna protein, light harvesting complex 2 (LH2) from Rhodopsuedomonas acidophila at the single-protein level. We utilize a novel single-molecule technique, the Anti-Brownian ELectrokinetic trap, to study LH2 in a solution-phase (non-perturbative) environment. By performing the first simultaneous measurements of fluorescence intensity, lifetime and spectra of single LH2 complexes, we identify three distinct states, and observe transitions occurring between them on a timescale of seconds. Our results reveal LH2 complexes undergo photoactivated switching to a quenched state, likely by a conformational change, and thermally revert to the ground state. This is a previously unknown, reversible quenching pathway, and is one mechanism through which photosynthetic organisms are able to adapt to changes in light intensities. Photosynthetic organisms flourish under low light intensities by converting photoenergy to chemical energy with near unity quantum efficiency and under high light intensities by safely dissipating excess photoenergy and deleterious photoproducts. The molecular mechanisms balancing these two functions remains incompletely described. One critical barrier to characterizing the mechanisms responsible for these processes is that they occur within proteins whose excited-state properties vary drastically between individual proteins and even within a single protein over time. In ensemble measurements, these excited-state properties appear only as the average value. To overcome this averaging, we investigate the purple bacterial antenna protein, light harvesting complex 2 (LH2) from Rhodopsuedomonas acidophila at the single-protein level. We utilize a novel single-molecule technique, the Anti-Brownian ELectrokinetic trap, to study LH2 in a solution-phase (non-perturbative) environment. By performing the first simultaneous measurements of fluorescence intensity, lifetime and spectra of single LH2 complexes, we identify three distinct states, and observe transitions occurring between them on a timescale of seconds. Our results reveal LH2 complexes undergo photoactivated switching to a quenched state, likely by a conformational change, and thermally revert to the ground state. This is a previously unknown, reversible quenching pathway, and is one mechanism through which photosynthetic organisms are able to adapt to changes in light intensities.
G.S. Schlau-Cohen, Q. Wang, J. Southall, R.J. Cogdell, W.E. Moerner, "Single-molecule spectroscopy reveals LH2 complexes switch between emissive states," Proc. Nat. Acad. Sci. (USA) 110, 10899-10903 (2013), published online 19 June 2013. DOI [ Slide] Slide]
Lifetime and spectrally resolved characterization of the photodynamics of SINGLE FLUOROPHORES in solution using the Anti-Brownian Electrokinetic trap
 We report simultaneous determination of fluorescence intensity, lifetime and emission spectrum over time scales on the order of seconds for single small organic molecules in solution, using the advanced recent design of the Anti-Brownian Electrokinetic trap. We demonstrate the technique with trapped single fluorophores of Atto647N and Alexa647. Three emission states with distinct intensities, lifetimes and emission peaks are found in the case of Atto647N. Transitions between states happen occasionally. We characterize the three states and quantify the transition probabilities between states using concurrent intensity, lifetime and spectrum data. Alexa647, on the other hand, showed little dynamics. These results represent a significant advance in the ability to identify and characterize different dynamical states of single molecules in aqueous solution with high precision and ms time resolution. We report simultaneous determination of fluorescence intensity, lifetime and emission spectrum over time scales on the order of seconds for single small organic molecules in solution, using the advanced recent design of the Anti-Brownian Electrokinetic trap. We demonstrate the technique with trapped single fluorophores of Atto647N and Alexa647. Three emission states with distinct intensities, lifetimes and emission peaks are found in the case of Atto647N. Transitions between states happen occasionally. We characterize the three states and quantify the transition probabilities between states using concurrent intensity, lifetime and spectrum data. Alexa647, on the other hand, showed little dynamics. These results represent a significant advance in the ability to identify and characterize different dynamical states of single molecules in aqueous solution with high precision and ms time resolution.
Quan Wang and W. E. Moerner, “Lifetime and spectrally resolved characterization of the photodynamics of single fluorophores in solution using the Anti-Brownian Electrokinetic trap,” Special Issue in memory of Paul F. Barbara, J. Phys. Chem. B 117, 4641-4648 (2013), published online 30 November 2012. DOI[ Slide] Slide]
Spectrally Resolved Anti-Brownian ELectrokinetic (ABEL) Trapping of Single Peridinin-Chlorophyll-Proteins in Solution
 We use an Anti-Brownian ELectrokinetic (ABEL) trap to probe spectral emission shifts in solution-phase single Peridinin-Chlorophyll-Proteins (PCPs). The ABEL trap allows localization of single biomolecules in solution in a small volume for extended observation without immobilization. The essential idea combines fluorescence-based position estimation with fast electrokinetic feedback in a microfluidic geometry to counter the Brownian motion of a single nanoscale object, hence maintaining its position in a sub-micron-sized field of view for hundreds of milliseconds to seconds. Peridinin-chlorophyll-protein is a water-soluble antenna protein found in dinoflagellates which uses peridinins (carotenoids) as accessory light harvesting pigments to absorb sunlight in the green region of the spectrum before transferring electronic excitation to chlorophyll. PCP is simpler than many other antenna complexes in that there are only two chlorophyll pigments per monomer which do not form an exciton. We use the ABEL trap to study single PCP monomers in solution for several seconds each. A significant fraction of the molecules show slow spectral shifts between two detection channels, red and blue in the figure (spectral diffusion) which can be characterized with one variable shown in black, relative to the bulk PCP spectrum. This is the first spectral emission measurement conducted in the ABEL trap. We use an Anti-Brownian ELectrokinetic (ABEL) trap to probe spectral emission shifts in solution-phase single Peridinin-Chlorophyll-Proteins (PCPs). The ABEL trap allows localization of single biomolecules in solution in a small volume for extended observation without immobilization. The essential idea combines fluorescence-based position estimation with fast electrokinetic feedback in a microfluidic geometry to counter the Brownian motion of a single nanoscale object, hence maintaining its position in a sub-micron-sized field of view for hundreds of milliseconds to seconds. Peridinin-chlorophyll-protein is a water-soluble antenna protein found in dinoflagellates which uses peridinins (carotenoids) as accessory light harvesting pigments to absorb sunlight in the green region of the spectrum before transferring electronic excitation to chlorophyll. PCP is simpler than many other antenna complexes in that there are only two chlorophyll pigments per monomer which do not form an exciton. We use the ABEL trap to study single PCP monomers in solution for several seconds each. A significant fraction of the molecules show slow spectral shifts between two detection channels, red and blue in the figure (spectral diffusion) which can be characterized with one variable shown in black, relative to the bulk PCP spectrum. This is the first spectral emission measurement conducted in the ABEL trap.
Samuel Bockenhauer, Quan Wang, and W. E. Moerner, “Spectrally Resolved Anti-Brownian ELectrokinetic (ABEL) Trapping of Single Peridinin-Chlorophyll-Proteins in Solution,” Proc. SPIE 8427, 84274C(1-9) (2012). DOI [ Slide] Slide]
Also see: S. Bockenhauer and W. E. Moerner, “Photo-Induced Conformational Flexibility in Single Solution-Phase Peridinin-Chlorophyll-Proteins,” J. Phys. Chem. A 117, 8399-8406 (2013),, published online 6 August 2013. DOI
Redox Cycling and Kinetic Analysis of Single Molecules of Solution-Phase Nitrite Reductase
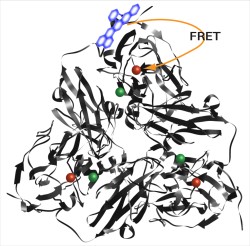 Single-molecule measurements are a valuable tool for revealing details of enzyme mechanisms by enabling observation of unsynchronized behavior. However, this approach often requires immobilizing the enzyme on a substrate, a process which may alter enzyme behavior. We apply a microfluidic trapping device to allow, for the first time, prolonged solution-phase measurement of single enzymes in solution. The redox state is sensed by a fluorescent dye attached to the protein near the type I Cu center so that when the Cu site is oxidized, the dye emission is partly quenched, but when the Cu site is reduced, the emission from the dye is unquenched. Individual redox cycling events are observed for single molecules of a blue nitrite reductase and are used to extract the microscopic kinetic parameters of the proposed catalytic cycle. Changes in parameters as a function of substrate concentration are consistent with a random-sequential substrate binding mechanism. Single-molecule measurements are a valuable tool for revealing details of enzyme mechanisms by enabling observation of unsynchronized behavior. However, this approach often requires immobilizing the enzyme on a substrate, a process which may alter enzyme behavior. We apply a microfluidic trapping device to allow, for the first time, prolonged solution-phase measurement of single enzymes in solution. The redox state is sensed by a fluorescent dye attached to the protein near the type I Cu center so that when the Cu site is oxidized, the dye emission is partly quenched, but when the Cu site is reduced, the emission from the dye is unquenched. Individual redox cycling events are observed for single molecules of a blue nitrite reductase and are used to extract the microscopic kinetic parameters of the proposed catalytic cycle. Changes in parameters as a function of substrate concentration are consistent with a random-sequential substrate binding mechanism.
Randall H. Goldsmith, Leandro C. Tabares, Dorota Kostrz, Christopher Dennison, Thijs J. Aartsma, Gerard W. Canters, and W. E. Moerner, “Redox cycling and kinetic analysis of single molecules of solution-phase nitrite reductase,” Proc. Nat. Acad. Sci. (USA) 108, 17269-17274 (2011), published online 3 October 2011. [ Slide] [Journal Link] Slide] [Journal Link]
Conformational Dynamics of Single G Protein-Coupled Receptors in Solution
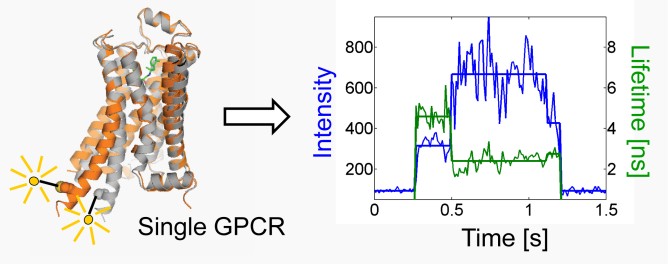 G Protein-Coupled Receptors (GPCRs) comprise a large family of seven-helix transmembrane proteins which regulate cellular signaling by sensing light, ligands, and binding proteins. The GPCR activation process, however, is not a simple on-off switch; current models suggest a complex conformational landscape in which the active, signaling state includes multiple conformations with similar downstream activity. The present study probes the conformational dynamics of single β2-Adrenergic Receptors (β2ARs) in the solution phase by Anti-Brownian ELectrokinetic (ABEL) trapping. The ABEL trap uses fast electrokinetic feedback in a microfluidic configuration to allow direct observation of a single fluorescently-labeled β2AR for hundreds of milliseconds to seconds. By choosing a reporter dye and labeling site sensitive to ligand binding, we observe a diversity of discrete fluorescence intensity and lifetime levels in single β2ARs, indicating a varying radiative lifetime and a range of discrete conformational states with dwell times of hundreds of milliseconds. We find that binding of agonist increases the dwell times of these states, and furthermore, we observe millisecond fluctuations within states. The intensity autocorrelations of these faster fluctuations are well-described by stretched exponential functions with stretching exponent β ~ 0.5, suggesting protein dynamics over a range of timescales. G Protein-Coupled Receptors (GPCRs) comprise a large family of seven-helix transmembrane proteins which regulate cellular signaling by sensing light, ligands, and binding proteins. The GPCR activation process, however, is not a simple on-off switch; current models suggest a complex conformational landscape in which the active, signaling state includes multiple conformations with similar downstream activity. The present study probes the conformational dynamics of single β2-Adrenergic Receptors (β2ARs) in the solution phase by Anti-Brownian ELectrokinetic (ABEL) trapping. The ABEL trap uses fast electrokinetic feedback in a microfluidic configuration to allow direct observation of a single fluorescently-labeled β2AR for hundreds of milliseconds to seconds. By choosing a reporter dye and labeling site sensitive to ligand binding, we observe a diversity of discrete fluorescence intensity and lifetime levels in single β2ARs, indicating a varying radiative lifetime and a range of discrete conformational states with dwell times of hundreds of milliseconds. We find that binding of agonist increases the dwell times of these states, and furthermore, we observe millisecond fluctuations within states. The intensity autocorrelations of these faster fluctuations are well-described by stretched exponential functions with stretching exponent β ~ 0.5, suggesting protein dynamics over a range of timescales.
Samuel Bockenhauer*, Alexandre Fürstenberg*, Xiao Jie Yao, Brian K. Kobilka, and W. E. Moerner (*equal contributions), "Conformational Dynamics of Single G Protein-Coupled Receptors in Solution," J. Phys. Chem. B 115, 13328-13338 (2011), published online 19 September 2011. [ Slide] [Journal Link] Slide] [Journal Link]
Sensing Cooperativity in ATP Hydrolysis for Single Multi-Subunit Enzymes in Solution
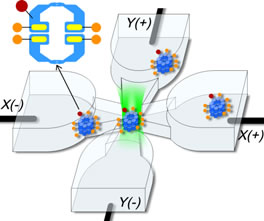 In order to operate in a synchronized fashion, multi-subunit enzymes use cooperative interactions intrinsic to their enzymatic cycle, but this process remains poorly understood. Accordingly, ATP number distributions in various hydrolyzed states have been obtained for single copies of the mammalian double-ring multi-subunit chaperonin TRiC/CCT in free solution using the emission from chaperonin-bound fluorescent nucleotides and closed-loop feedback trapping provided by an Anti-Brownian ELectrokinetic trap (ABEL trap). Observations of the 16-subunit complexes as ADP molecules are dissociating shows a peak in the bound ADP number distribution at 8 ADP, whose height falls over time with little shift in the position of the peak, indicating a highly cooperative ADP release process which would be difficult to observe by ensemble-averaged methods. When AlFx is added to produce ATP hydrolysis transition state mimics (ADP∙AlFx) locked to the complex, the peak at 8 nucleotides dominates for all but the lowest incubation concentrations. Although ensemble averages of the single-molecule data show agreement with standard cooperativity models, surprisingly, the observed number distributions depart from standard models, illustrating the value of these single-molecule observations in constraining the mechanism of cooperativity. While a complete alternative microscopic model cannot be defined at present, the addition of subunit-occupancy-dependent cooperativity in hydrolysis yields distributions consistent with the data. In order to operate in a synchronized fashion, multi-subunit enzymes use cooperative interactions intrinsic to their enzymatic cycle, but this process remains poorly understood. Accordingly, ATP number distributions in various hydrolyzed states have been obtained for single copies of the mammalian double-ring multi-subunit chaperonin TRiC/CCT in free solution using the emission from chaperonin-bound fluorescent nucleotides and closed-loop feedback trapping provided by an Anti-Brownian ELectrokinetic trap (ABEL trap). Observations of the 16-subunit complexes as ADP molecules are dissociating shows a peak in the bound ADP number distribution at 8 ADP, whose height falls over time with little shift in the position of the peak, indicating a highly cooperative ADP release process which would be difficult to observe by ensemble-averaged methods. When AlFx is added to produce ATP hydrolysis transition state mimics (ADP∙AlFx) locked to the complex, the peak at 8 nucleotides dominates for all but the lowest incubation concentrations. Although ensemble averages of the single-molecule data show agreement with standard cooperativity models, surprisingly, the observed number distributions depart from standard models, illustrating the value of these single-molecule observations in constraining the mechanism of cooperativity. While a complete alternative microscopic model cannot be defined at present, the addition of subunit-occupancy-dependent cooperativity in hydrolysis yields distributions consistent with the data.
Yan Jiang, Nicholai R. Douglas, Nicholas R. Conley, Erik J. Miller, Judith Frydman, and W. E. Moerner, “Sensing Cooperativity in ATP Hydrolysis for Single Multi-Subunit Enzymes in Solution,” Proc. Nat. Acad. Sci. (USA) 108, 16962-16967 (2011), published online 6 September 2011. [ Slide] [Journal Link][Commentary] Slide] [Journal Link][Commentary]
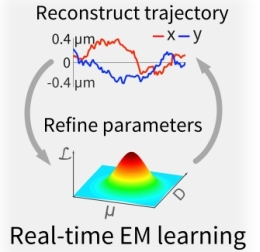
An Adaptive Anti-Brownian ELectrokinetic (ABEL) Trap with Real-time Information on Single-Molecule Diffusivity and Mobility
 We present the design and implementation of an adaptive Anti-Brownian ELectrokinetic (ABEL) trap capable of extracting estimates of the diffusion coefficient and mobility of single trapped fluorescent nanoscale objects such as biomolecules in solution. This represents an experimental implementation of the optimal trapping strategy published earlier (see below). The system features rapid acousto-optic scanning of a confocal excitation spot on a 2D square lattice in the path of a Knight's tour to encode position information on the arrival time of each detected photon, and a Kalman filter-based signal processing unit for refined position estimation. We demonstrate stable trapping of multi-subunit proteins (D ~ 22 µm2/s ) with a count rate of 6 kHz for as long as 15 seconds and small single-stranded DNA molecules (D ~ 118 µm2/s) at a 15 kHz count rate for seconds. Moreover, we demonstrate real-time measurement of diffusion coefficient and electrokinetic mobility of trapped objects, using adaptive tuning of the Kalman filter parameters. We present the design and implementation of an adaptive Anti-Brownian ELectrokinetic (ABEL) trap capable of extracting estimates of the diffusion coefficient and mobility of single trapped fluorescent nanoscale objects such as biomolecules in solution. This represents an experimental implementation of the optimal trapping strategy published earlier (see below). The system features rapid acousto-optic scanning of a confocal excitation spot on a 2D square lattice in the path of a Knight's tour to encode position information on the arrival time of each detected photon, and a Kalman filter-based signal processing unit for refined position estimation. We demonstrate stable trapping of multi-subunit proteins (D ~ 22 µm2/s ) with a count rate of 6 kHz for as long as 15 seconds and small single-stranded DNA molecules (D ~ 118 µm2/s) at a 15 kHz count rate for seconds. Moreover, we demonstrate real-time measurement of diffusion coefficient and electrokinetic mobility of trapped objects, using adaptive tuning of the Kalman filter parameters.
Quan Wang and W. E. Moerner, "An Adaptive Anti-Brownian ELectrokinetic (ABEL) Trap with Real-time Information on Single-Molecule Diffusivity and Mobility," ACS Nano 5, 5792-5799 (2011) published online May 25, 2011. [ Slide] [Journal Link] Slide] [Journal Link]
Watching conformational- and photodynamics of single fluorescent proteins in solution
 Observing the dynamics of single
biomolecules over prolonged time periods is difficult to achieve without significantly altering the molecule
through immobilization. It can, however, be accomplished using the Anti-Brownian ELectrokinetic (ABEL) Trap,
which allows extended investigation of solution-phase biomolecules - without immobilization - through
real-time electrokinetic feedback. Here we apply the ABEL trap to study an important photosynthetic antenna
protein, Allophycocyanin (APC). The technique allows the observation of single molecules of solution-phase APC for
more than one second. We observe a complex relationship between fluorescence intensity and lifetime that cannot be
explained by simple static kinetic models. Light-induced conformational changes are shown to occur and evidence is
obtained for fluctuations in the spontaneous emission lifetime, which is typically assumed to be constant. Our methods
provide a new window into the dynamics of fluorescent proteins and the observations are relevant for the
interpretation of in vivo single-molecule imaging experiments, bacterial photosynthetic regulation, and biomaterials
for solar energy harvesting. Observing the dynamics of single
biomolecules over prolonged time periods is difficult to achieve without significantly altering the molecule
through immobilization. It can, however, be accomplished using the Anti-Brownian ELectrokinetic (ABEL) Trap,
which allows extended investigation of solution-phase biomolecules - without immobilization - through
real-time electrokinetic feedback. Here we apply the ABEL trap to study an important photosynthetic antenna
protein, Allophycocyanin (APC). The technique allows the observation of single molecules of solution-phase APC for
more than one second. We observe a complex relationship between fluorescence intensity and lifetime that cannot be
explained by simple static kinetic models. Light-induced conformational changes are shown to occur and evidence is
obtained for fluctuations in the spontaneous emission lifetime, which is typically assumed to be constant. Our methods
provide a new window into the dynamics of fluorescent proteins and the observations are relevant for the
interpretation of in vivo single-molecule imaging experiments, bacterial photosynthetic regulation, and biomaterials
for solar energy harvesting.
Randall H. Goldsmith and W. E. Moerner, "Watching conformational- and photo-dynamics of single fluorescent
proteins in solution," Nature Chemistry 2,179-186 (2010) published online January 31, 2010. [ Slide] [Journal Link] [News and Views] Slide] [Journal Link] [News and Views]
Optimal Strategy for Trapping Single Fluorescent Molecules in Solution Using the ABEL Trap
 Trapping of 10-nm-sized single fluorescent
bio-molecules in solution has been achieved using
high-speed position sensing and electrokinetic feedback forces in the Anti-Brownian ELectrokinetic (ABEL) trap.
The high diffusion coefficient of small objects in solution requires very fast, real-time sensing of position,
and this has been previously achieved using a simple rotating beam, but improved strategies are needed for the smallest objects,
such as single nanometer-sized fluorescent molecules. At the same time, single molecules are limited in photon emission rate
and total number of photons, so each emitted photon must be used as efficiently as possible. We describe a new controller
design for the ABEL trap which features fast, knight's tour scanning of an excitation beam on a 2D square
lattice and a Kalman filter-based estimator for optimal position sensing. This strategy leads directly to a
maximum-likelihood-based method to extract the diffusion coefficient of the object held in the trap.
The effectiveness of the algorithms are demonstrated and compared to the simple rotating beam design through
Monte Carlo simulations. Our new approach yields tighter trapping and a much improved ability to extract
diffusion coefficients. Trapping of 10-nm-sized single fluorescent
bio-molecules in solution has been achieved using
high-speed position sensing and electrokinetic feedback forces in the Anti-Brownian ELectrokinetic (ABEL) trap.
The high diffusion coefficient of small objects in solution requires very fast, real-time sensing of position,
and this has been previously achieved using a simple rotating beam, but improved strategies are needed for the smallest objects,
such as single nanometer-sized fluorescent molecules. At the same time, single molecules are limited in photon emission rate
and total number of photons, so each emitted photon must be used as efficiently as possible. We describe a new controller
design for the ABEL trap which features fast, knight's tour scanning of an excitation beam on a 2D square
lattice and a Kalman filter-based estimator for optimal position sensing. This strategy leads directly to a
maximum-likelihood-based method to extract the diffusion coefficient of the object held in the trap.
The effectiveness of the algorithms are demonstrated and compared to the simple rotating beam design through
Monte Carlo simulations. Our new approach yields tighter trapping and a much improved ability to extract
diffusion coefficients.
Quan Wang and W. E. Moerner, "Optimal strategy for trapping single fluorescent molecules in solution using the ABEL trap," Appl. Phys. B 99, 23-30 (2010), published online December 12, 2009. [ Slide] [Journal Link] Slide] [Journal Link]
Dynamics of Single DNA Molecules in Equilibrium
 Thermal fluctuations agitate molecules in solution
over a broad range of times and distances. By passively watching the shape fluctuations of a thermally
driven biomolecule, one can infer properties of the underlying interactions that determine the motion.
We applied this concept to single molecules of fluorescently labeled λ-DNA, a key model system for
polymer physics. In contrast to most other single-molecule DNA experiments, we examined the unstretched,
equilibrium state of DNA by using an anti-Brownian electrokinetic trap to confine the center of mass of the DNA
without perturbing its internal dynamics. We analyze the long-wavelength conformational normal modes, calculate
their spring constants, and measure linear and nonlinear couplings between modes. The modes show strong signs of
nonlinear hydrodynamics, a feature of the underlying equations of polymer dynamics that has not previously been
reported and is neglected in the widely used Rouse and Zimm approximations. Thermal fluctuations agitate molecules in solution
over a broad range of times and distances. By passively watching the shape fluctuations of a thermally
driven biomolecule, one can infer properties of the underlying interactions that determine the motion.
We applied this concept to single molecules of fluorescently labeled λ-DNA, a key model system for
polymer physics. In contrast to most other single-molecule DNA experiments, we examined the unstretched,
equilibrium state of DNA by using an anti-Brownian electrokinetic trap to confine the center of mass of the DNA
without perturbing its internal dynamics. We analyze the long-wavelength conformational normal modes, calculate
their spring constants, and measure linear and nonlinear couplings between modes. The modes show strong signs of
nonlinear hydrodynamics, a feature of the underlying equations of polymer dynamics that has not previously been
reported and is neglected in the widely used Rouse and Zimm approximations.
A. E. Cohen and W. E. Moerner, "Dynamics of Single DNA Molecules in Equilibrium," Proc. Nat. Acad. Sci. (USA) 104, 12596-12602 (published online May 11, 2007) [PDF] [Movie] [Supporting Information] [Journal Link]
Also see: Conformations of a Single DNA Molecule in Equilibrium
Press:
Original ABEL Trap Publications
High speed hardware version of the ABEL trap: Controlling Brownian Motion of Single Proteins and Single Fluorophores in Aqueous Buffer, Optics Express 16, 6941-6956 (2008) [PDF]
Internal Mechanical Response of a Polymer in Solution, Phys. Rev. Lett. 98, 116001-(1-4) (2007)
[PDF] [Supporting Information]
Suppressing Brownian Motion of Individual Biomolecules in Solution (Proc. Nat. Acad. Sci. (USA) v. 103, 4362 (2006)) [PDF] [Journal link] [Supporting Material and Movies].
An All-Glass Microfluidic Cell for the ABEL Trap: Fabrication and Modeling (Proc. SPIE v. 5930, 59300S-1-S-8 (2005)) [PDF]
Control of Nanoparticles with Arbitrary Two-Dimensional Force Fields (AEC, Phys. Rev. Lett. v. 94, 188102 (2005)) [PDF]
The Anti-Brownian ELectrophoretic Trap: Fabrication and Software (Proc. SPIE v. 5699, pp. 296-305 (2005))
[PDF]
Method for trapping and manipulating
nanoscale objects in solution (Appl. Phys. Lett. v. 86, 093109
(2005)) [Slide]
[PDF]
|