Multiple Linear Regression Inference#
Download#
Outline#
Case studies:
A. Galileo’s falling bodies
B. Energy costs of echolocation
Energy costs of echolocation#
url = 'https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/bats_energy.csv'
bats = read.table(url, sep=',', header=TRUE)
bats
| Mass | Type | Energy |
|---|---|---|
| <dbl> | <chr> | <dbl> |
| 779.0 | non-echolocating bats | 43.70 |
| 628.0 | non-echolocating bats | 34.80 |
| 258.0 | non-echolocating bats | 23.30 |
| 315.0 | non-echolocating bats | 22.40 |
| 24.3 | non-echolocating birds | 2.46 |
| 35.0 | non-echolocating birds | 3.93 |
| 72.8 | non-echolocating birds | 9.15 |
| 120.0 | non-echolocating birds | 13.80 |
| 213.0 | non-echolocating birds | 14.60 |
| 275.0 | non-echolocating birds | 22.80 |
| 370.0 | non-echolocating birds | 26.20 |
| 384.0 | non-echolocating birds | 25.90 |
| 442.0 | non-echolocating birds | 29.50 |
| 412.0 | non-echolocating birds | 43.70 |
| 330.0 | non-echolocating birds | 34.00 |
| 480.0 | non-echolocating birds | 27.80 |
| 93.0 | echolocating bats | 8.83 |
| 8.0 | echolocating bats | 1.35 |
| 6.7 | echolocating bats | 1.12 |
| 7.7 | echolocating bats | 1.02 |
Fitting a model#
bats.lm = lm(Energy ~ Type + Mass, data=bats)
summary(bats.lm)
Call:
lm(formula = Energy ~ Type + Mass, data = bats)
Residuals:
Min 1Q Median 3Q Max
-5.8198 -3.6711 -0.9508 1.1499 13.9899
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.421257 2.660684 0.534 0.601
Typenon-echolocating bats 1.168512 5.145112 0.227 0.823
Typenon-echolocating birds 4.600720 3.537113 1.301 0.212
Mass 0.057495 0.007557 7.608 1.06e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 5.303 on 16 degrees of freedom
Multiple R-squared: 0.8791, Adjusted R-squared: 0.8565
F-statistic: 38.79 on 3 and 16 DF, p-value: 1.437e-07
Other models#
simpler.lm = lm(Energy ~ Type, data=bats)
coef(simpler.lm)
- (Intercept)
- 3.07999999999999
- Typenon-echolocating bats
- 27.97
- Typenon-echolocating birds
- 18.0733333333333
complex.lm = lm(Energy ~ Type + Mass + Type:Mass, data=bats)
coef(complex.lm)
- (Intercept)
- 0.493983042663024
- Typenon-echolocating bats
- 10.7334006739125
- Typenon-echolocating birds
- 2.82275854753212
- Mass
- 0.0896366363028415
- Typenon-echolocating bats:Mass
- -0.0495909468413778
- Typenon-echolocating birds:Mass
- -0.021861993030729
Case study B: Galileo’s data#
url = 'https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/galileo.csv'
galileo = read.table(url, sep=',', header=TRUE)
with(galileo, plot(Height, Distance))
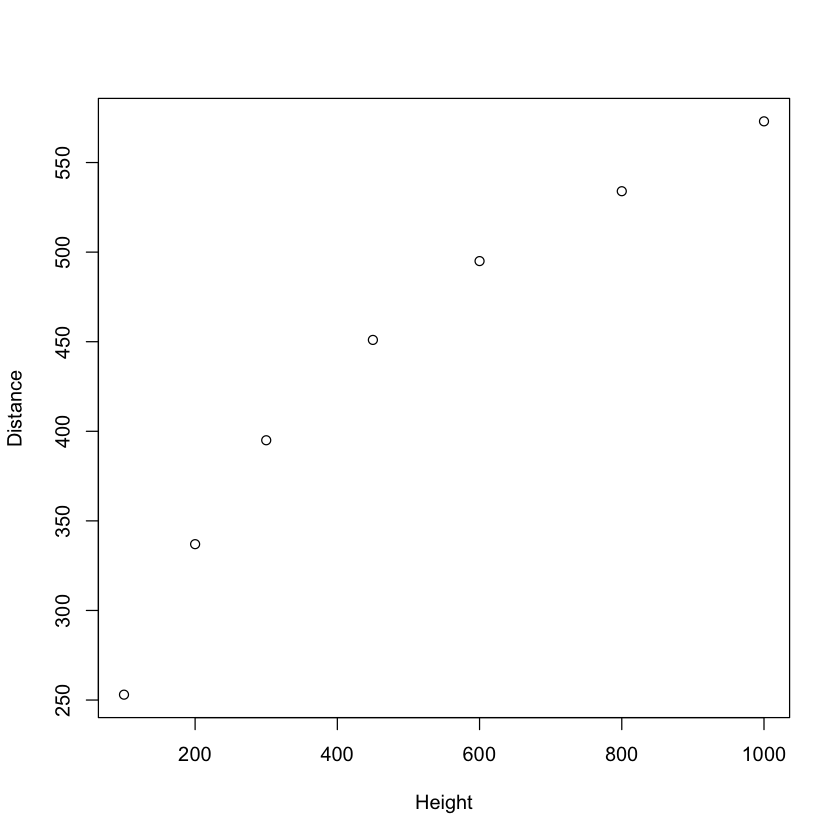
Galileo fit a quadratic model to his data
Note the notation
I(Height^2)– withoutIa quadratic term will not be added…
galileo1.lm = lm(Distance ~ Height + I(Height^2), data=galileo)
summary(galileo1.lm)
Call:
lm(formula = Distance ~ Height + I(Height^2), data = galileo)
Residuals:
1 2 3 4 5 6 7
-14.308 9.170 13.523 1.940 -6.177 -12.607 8.458
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.999e+02 1.676e+01 11.928 0.000283 ***
Height 7.083e-01 7.482e-02 9.467 0.000695 ***
I(Height^2) -3.437e-04 6.678e-05 -5.147 0.006760 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 13.64 on 4 degrees of freedom
Multiple R-squared: 0.9903, Adjusted R-squared: 0.9855
F-statistic: 205 on 2 and 4 DF, p-value: 9.333e-05
A different way to fit the model
galileo2.lm = lm(Distance ~ poly(Height, 2), data=galileo)
summary(galileo2.lm)
Call:
lm(formula = Distance ~ poly(Height, 2), data = galileo)
Residuals:
1 2 3 4 5 6 7
-14.308 9.170 13.523 1.940 -6.177 -12.607 8.458
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 434.000 5.155 84.190 1.19e-07 ***
poly(Height, 2)1 267.116 13.639 19.585 4.01e-05 ***
poly(Height, 2)2 -70.194 13.639 -5.147 0.00676 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 13.64 on 4 degrees of freedom
Multiple R-squared: 0.9903, Adjusted R-squared: 0.9855
F-statistic: 205 on 2 and 4 DF, p-value: 9.333e-05
Predictions / CIs are the same
predict(galileo1.lm, list(Height=250), interval='confidence')
predict(galileo2.lm, list(Height=250), interval='confidence')
| fit | lwr | upr | |
|---|---|---|---|
| 1 | 355.5126 | 337.1189 | 373.9063 |
| fit | lwr | upr | |
|---|---|---|---|
| 1 | 355.5126 | 337.1189 | 373.9063 |
Tests of quadratic effect are the same
galileo0.lm = lm(Distance ~ Height, data=galileo)
anova(galileo0.lm, galileo1.lm)
anova(galileo0.lm, galileo2.lm)
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 5 | 5671.2063 | NA | NA | NA | NA |
| 2 | 4 | 744.0781 | 1 | 4927.128 | 26.48716 | 0.006760485 |
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 5 | 5671.2063 | NA | NA | NA | NA |
| 2 | 4 | 744.0781 | 1 | 4927.128 | 26.48716 | 0.006760485 |
Confidence intervals#
Suppose we want a \((1-\alpha)\cdot 100\%\) CI for \(\sum_{j=0}^p a_j\beta_j\).
Just as in simple linear regression:
\[\sum_{j=0}^p a_j \widehat{\beta}_j \pm t_{1-\alpha/2, n-p-1} \cdot SE\left(\sum_{j=0}^p a_j\widehat{\beta}_j\right).\]
confint(bats.lm, level=0.90)
predict(bats.lm, list(Mass=300, Type="non-echolocating bats"), interval='confidence')
| 5 % | 95 % | |
|---|---|---|
| (Intercept) | -3.2239868 | 6.06650127 |
| Typenon-echolocating bats | -7.8142562 | 10.15127927 |
| Typenon-echolocating birds | -1.5746672 | 10.77610686 |
| Mass | 0.0443021 | 0.07068873 |
| fit | lwr | upr | |
|---|---|---|---|
| 1 | 19.83839 | 13.40731 | 26.26948 |
\(T\)-statistics revisited#
Of course, these confidence intervals are based on the standard ingredients of a \(T\)-statistic.
Suppose we want to test
As in simple linear regression, it is based on
If \(H_0\) is true, then \(T \sim t_{n-p-1}\), so we reject \(H_0\) at level \(\alpha\) if
Let’s do a quick calculation to remind ourselves the relationships of the variables in the table above.
T_Mass = 0.057495 / 0.007557
P_Mass = 2 * (1 - pt(abs(T_Mass), 16))
c(T_Mass, P_Mass)
- 7.60817784835252
- 1.05724836840793e-06
summary(bats.lm)$coef
| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 1.42125723 | 2.660683583 | 0.534170 | 6.005680e-01 |
| Typenon-echolocating bats | 1.16851155 | 5.145112379 | 0.227111 | 8.232139e-01 |
| Typenon-echolocating birds | 4.60071984 | 3.537112527 | 1.300699 | 2.117840e-01 |
| Mass | 0.05749542 | 0.007556812 | 7.608422 | 1.056817e-06 |
One-sided tests#
Suppose, instead, we wanted to test the one-sided hypothesis
If \(H_0\) is true, then \(T\) is no longer exactly \(t_{n-p-1}\) but we still have
We reject \(H_0\) at level \(\alpha\) if
Standard error of \(\sum_{j=0}^p a_j \widehat{\beta}_j\)#
In order to form these \(T\) statistics, we need the \(SE\) of our estimate \(\sum_{j=0}^p a_j \widehat{\beta}_j\).
Based on matrix approach to regression
Don’t worry too much about specific implementation – for much of the effects we want
Rwill do this for you in general.
X = model.matrix(bats.lm)
sigma.hat = sqrt(sum(resid(bats.lm)^2 / 16))
cov.beta = sigma.hat^2 * solve(t(X) %*% X)
cov.beta
| (Intercept) | Typenon-echolocating bats | Typenon-echolocating birds | Mass | |
|---|---|---|---|---|
| (Intercept) | 7.079237129 | -6.26372902 | -6.64565865 | -1.647491e-03 |
| Typenon-echolocating bats | -6.263729023 | 26.47218139 | 13.26936531 | -2.661969e-02 |
| Typenon-echolocating birds | -6.645658653 | 13.26936531 | 12.51116503 | -1.338123e-02 |
| Mass | -0.001647491 | -0.02661969 | -0.01338123 | 5.710541e-05 |
vcov(bats.lm)
| (Intercept) | Typenon-echolocating bats | Typenon-echolocating birds | Mass | |
|---|---|---|---|---|
| (Intercept) | 7.079237129 | -6.26372902 | -6.64565865 | -1.647491e-03 |
| Typenon-echolocating bats | -6.263729023 | 26.47218139 | 13.26936531 | -2.661969e-02 |
| Typenon-echolocating birds | -6.645658653 | 13.26936531 | 12.51116503 | -1.338123e-02 |
| Mass | -0.001647491 | -0.02661969 | -0.01338123 | 5.710541e-05 |
The standard errors of each coefficient estimate are the square root of the diagonal entries. They appear as the
Std. Error column in the coef table.
sqrt(diag(vcov(bats.lm)))
summary(bats.lm)$coef[,2]
- (Intercept)
- 2.66068358303546
- Typenon-echolocating bats
- 5.14511237921139
- Typenon-echolocating birds
- 3.53711252678651
- Mass
- 0.00755681226189849
- (Intercept)
- 2.66068358303546
- Typenon-echolocating bats
- 5.14511237921139
- Typenon-echolocating birds
- 3.53711252678651
- Mass
- 0.00755681226189849
Prediction / forecasting interval#
Basically identical to simple linear regression.
Prediction interval at \(X_{1,new}, \dots, X_{p,new}\):
Questions about many (combinations) of \(\beta_j\)’s#
In multiple regression we can ask more complicated questions than in simple regression.
For instance, in
bats.lmwe could ask whetherTypeis important at all?These questions can be answered answered by \(F\)-statistics.
Dropping one or more variables#
Suppose we wanted to test the above hypothesis Formally, the null hypothesis is:
The alternative is
This test is an \(F\)-test based on two models
\(SSE\) of a model#
In the graphic, a “model”, \({\cal M}\) is a subspace of \(\mathbb{R}^n\) (e.g. column space of \({X}\)).
Least squares fit = projection onto the subspace of \({\cal M}\), yielding predicted values \(\widehat{Y}_{{\cal M}}\)
Error sum of squares:
Least squares for \(F\) statistic#
 {height=400 fig-align=”center”}
{height=400 fig-align=”center”}
Fits of a full and reduced model \(\hat{Y}_F\) and \(\hat{Y}_R\)
The difference \(\hat{Y}_F-\hat{Y}_R\).
Right triangle for \(F\) statistic#
 {height=300 fig-align=”center”}
{height=300 fig-align=”center”}
Sides of the triangle: \(SSE_R-SSE_F\), \(SSE_F\)
Hypotenuse: \(SSE_R\)
Right triangle with full and reduced model: degrees of freedom#
 {height=300 fig-align=”center”}
{height=300 fig-align=”center”}
Sides of the triangle: \(df_R-df_F\), \(df_F\)
Hypotenuse: \(df_R\)
\(F\)-statistic for \(H_0:\beta_{1}=\beta_{2}=0\)#
We compute the \(F\) statistic the same to compare any two (nested) models
Reject \(H_0\) at level \(\alpha\) if \(F > F_{1-\alpha, 2, 16}\).
reduced.lm = lm(Energy ~ Mass, data=bats)
anova(reduced.lm, bats.lm)
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 18 | 528.8563 | NA | NA | NA | NA |
| 2 | 16 | 450.0292 | 2 | 78.82704 | 1.401278 | 0.2749315 |
General \(F\)-tests#
Given two models \(R \subset F\) (i.e. \(R\) is a subspace of \(F\)), we can consider testing
The least squares picture has models \(X_R\) and \(X_F=X_R+(X_F \perp X_R) \dots\)
The test statistic is
If \(H_0\) is true, \(F \sim F_{df_R-df_F, df_F}\) so we reject \(H_0\) at level \(\alpha\) if \(F > F_{1-\alpha, df_R-df_F, df_F}\).
Constraining coefficients#
Suppose we wanted to test \(H_0\): the line for non-echolocating bats has the same intercept as the line for non-echolocating birds.
Can be expressed as \(H_0:\beta_1=\beta_2\) in
bats.lm.
Strategy 1: fit a model in which this is forced to be true#
null_group = rep(1, nrow(bats))
null_group[bats$Type=="echolocating bats"] = 2
bats$null_group = factor(null_group)
null_bats.lm = lm(Energy ~ Mass + null_group, data=bats)
anova(null_bats.lm, bats.lm)
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 17 | 476.6541 | NA | NA | NA | NA |
| 2 | 16 | 450.0292 | 1 | 26.62481 | 0.9465984 | 0.345067 |
Strategy 2: a \(T\)-statistic#
Hypothesis is \(H_0:\beta_1-\beta_2=0\)
This method doesn’t require fitting the special model
null_bats.lm!Can be generalized to \(F\) tests (hypotheses involving multiple contrasts of \(\beta\))
A = c(0,1,-1,0)
var_diff = sum(A * vcov(bats.lm) %*% A)
se_diff = sqrt(var_diff)
T = sum(coef(bats.lm) * A) / se_diff
T^2
Math aside: general linear hypothesis#
Suppose we want to test the null hypothesis
Alternative is
Math aside: \(F\) statistic in general linear hypothesis#
Numerator
Denominator: the usual MSE
We just used special case \(q=1\) above…