Simple Linear Regression#
Download#
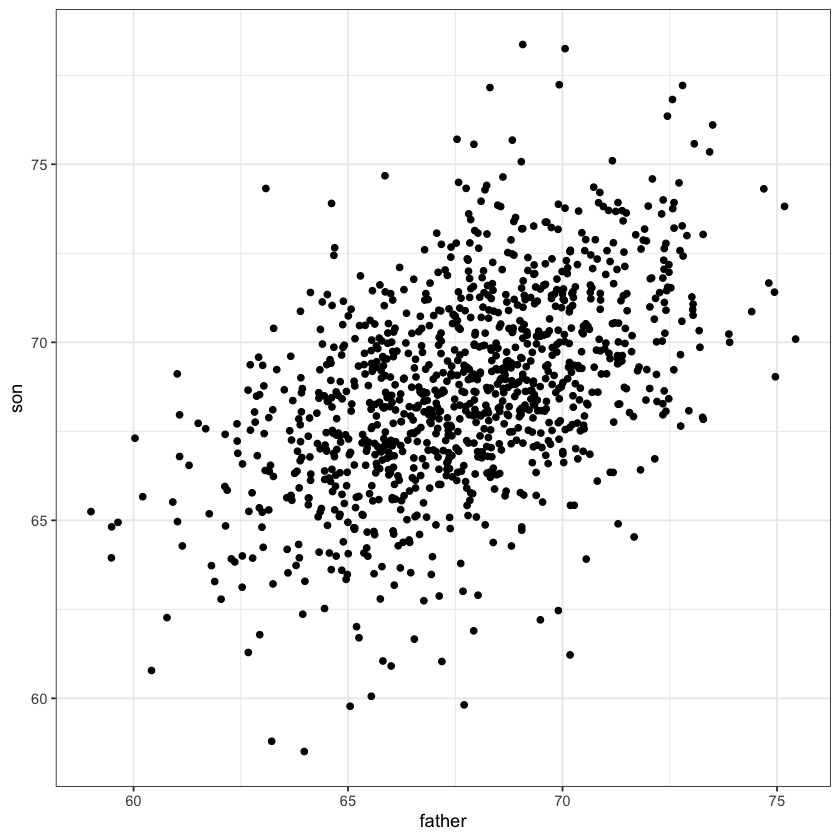
Father & son data#
Pearson’s height data#
library(UsingR)
heights = UsingR::father.son
head(heights)
Loading required package: MASS
Loading required package: HistData
Loading required package: Hmisc
Attaching package: ‘Hmisc’
The following objects are masked from ‘package:base’:
format.pval, units
| fheight | sheight | |
|---|---|---|
| <dbl> | <dbl> | |
| 1 | 65.04851 | 59.77827 |
| 2 | 63.25094 | 63.21404 |
| 3 | 64.95532 | 63.34242 |
| 4 | 65.75250 | 62.79238 |
| 5 | 61.13723 | 64.28113 |
| 6 | 63.02254 | 64.24221 |
# for scatter plots of heights
set.seed(1)
library(ggplot2)
library(UsingR)
heights = UsingR::father.son
heights$father = heights$fheight
heights$son = heights$sheight
height.lm = lm(son ~ father, data=heights)
intercept = height.lm$coef[1]
slope = height.lm$coef[2]
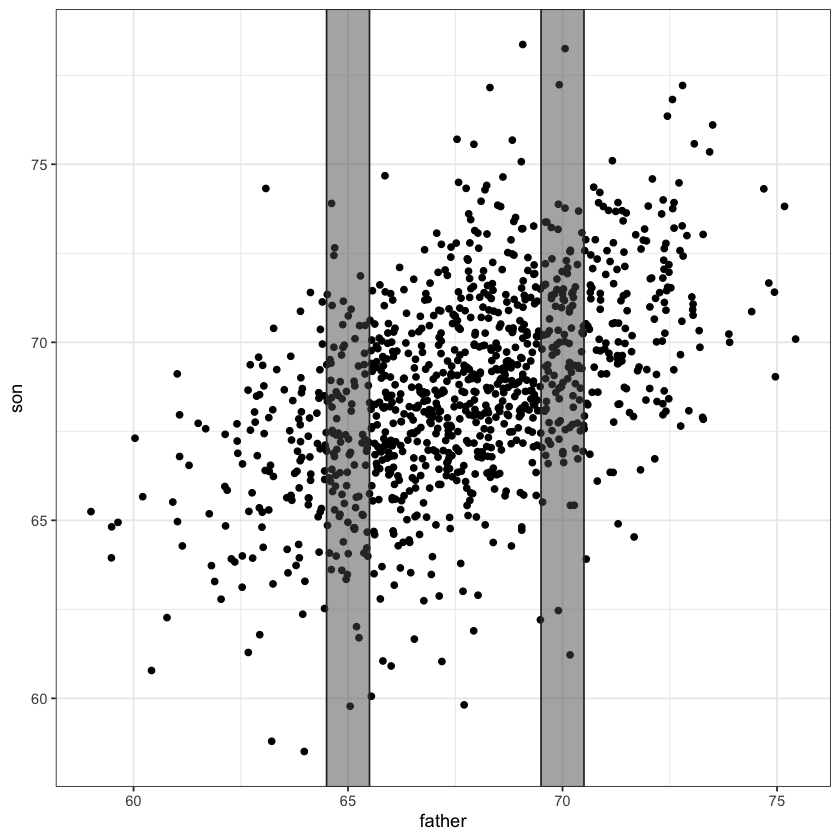
Father & son data#
library(ggplot2)
heights_fig <- ggplot(heights, aes(father, son)) + geom_point() + theme_bw()
heights_fig
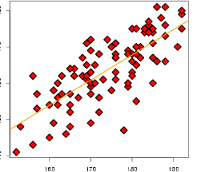
An example of simple linear regression model.
Breakdown of terms:
regression model: a model for the mean of a response given features
linear: model of the mean is linear in parameters of interest
simple: only a single feature
Slicewise model#
#| fig-align: center
rect = data.frame(xmin=69.5, xmax=70.5, ymin=-Inf, ymax=Inf)
heights_fig + geom_rect(data=rect, aes(xmin=xmin, xmax=xmax, ymin=ymin, ymax=ymax),
color="grey20",
alpha=0.5,
inherit.aes = FALSE)
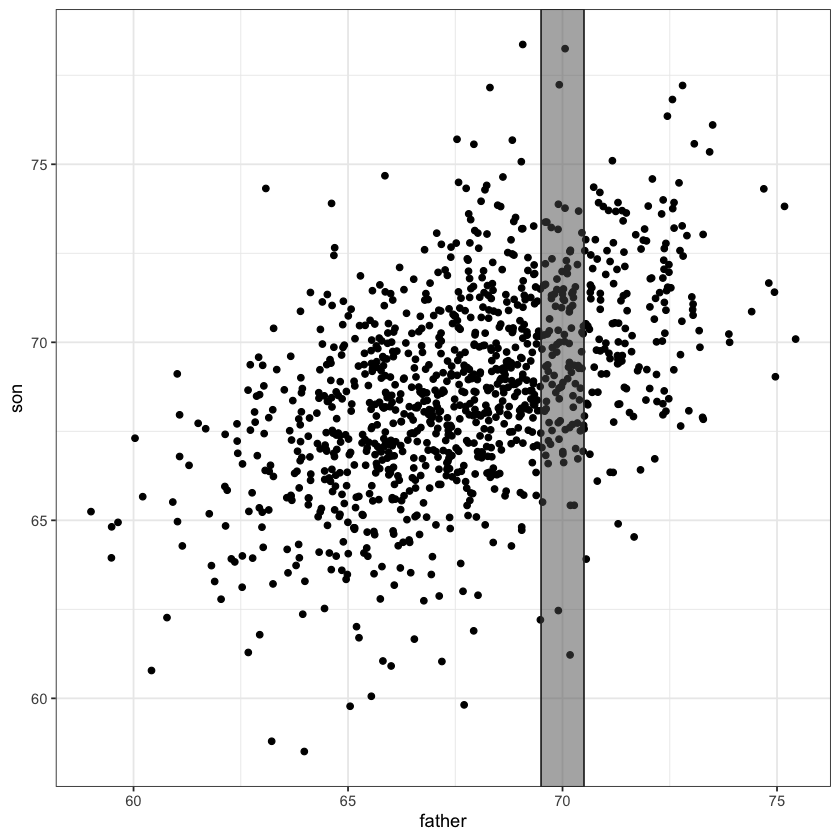
A simple linear regression model fits a line through the above scatter plot by modelling slices.
Conditional means#
father = heights$fheight
son = heights$sheight
selected_points = (father <= 70.5) & (father >= 69.5)
mean(son[selected_points])
The average height of sons whose height fell within our slice [69.5,79.5] is about 69.8 inches.
This height varies by slice…
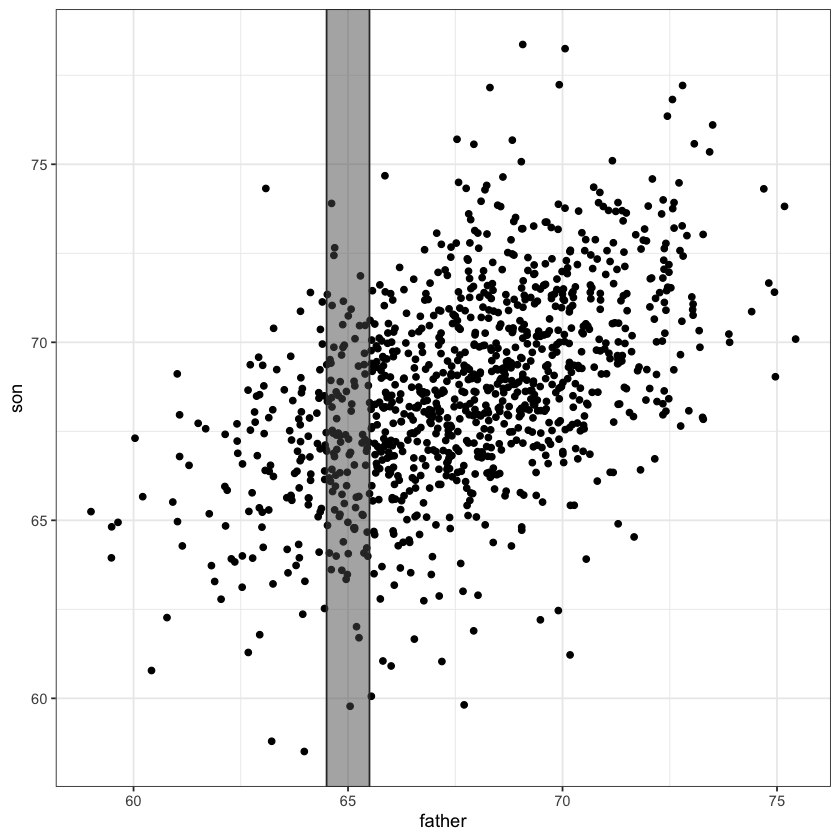
At 65 inches it’s about 67.2 inches:
selected_points = (father <= 65.5) & (father >= 64.5)
mean(son[selected_points])
rect = data.frame(xmin=64.5, xmax=65.5, ymin=-Inf, ymax=Inf)
heights_fig + geom_rect(data=rect, aes(xmin=xmin, xmax=xmax, ymin=ymin, ymax=ymax),
color="grey20",
alpha=0.5,
inherit.aes = FALSE)
Multiple samples model (longevity example)#
We’ve seen this slicewise model before: error in each slice the same.
longevity = read.csv('https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/longevity.csv', header=TRUE)
boxplot(Lifetime ~ Diet,
data=longevity,
col='orange',
pch=23,
bg='red')
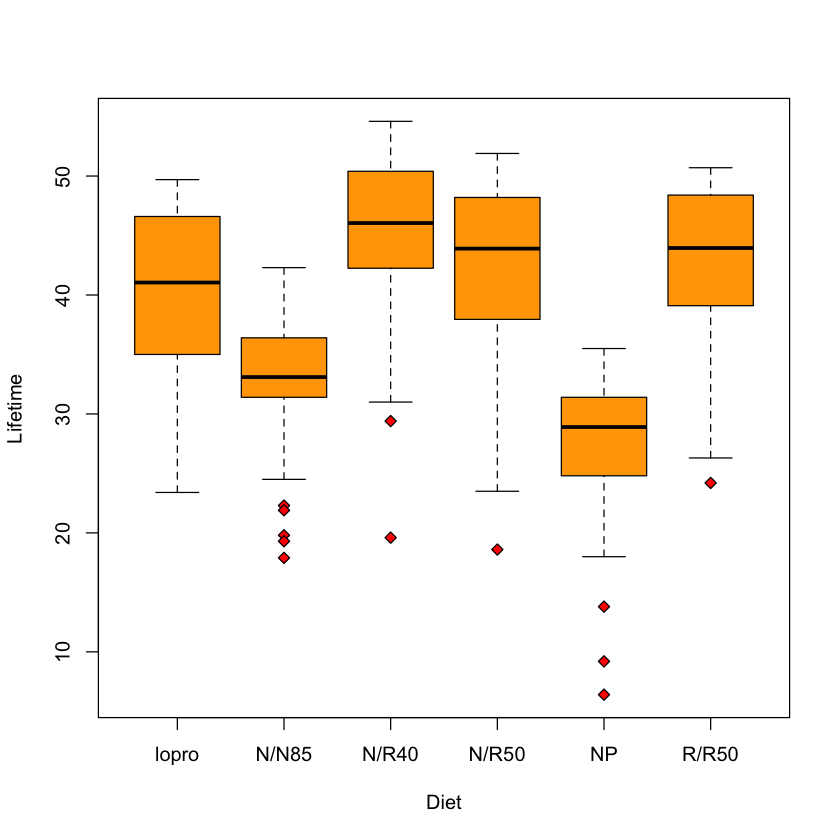
Regression as slicewise model#
In
longevitymodel: no relation between the means in each slice! We needed to use a parameter for eachDiet…Regression model says that the mean in slice
fatheris
This ties together all
(father, son)points in the scatterplot.Chooses \((\beta_0, \beta_1)\) by jointly modeling the mean in each slice…
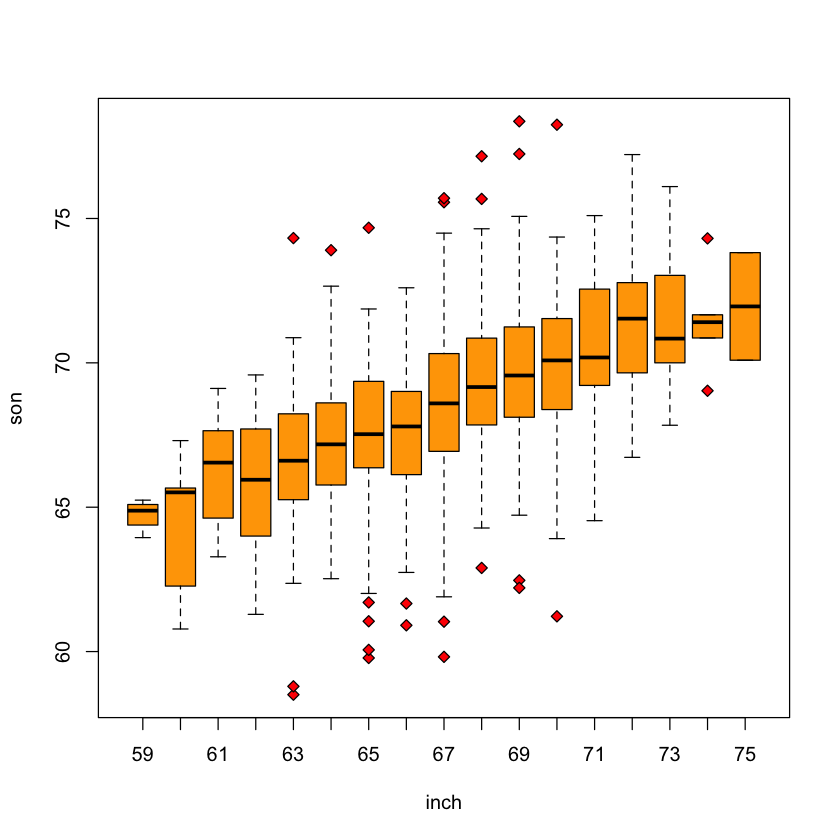
Height data as slices#
inch = factor(as.integer(father))
boxplot(son ~ inch,
col='orange',
pch=23,
bg='red')
heights_fig + geom_abline(slope=slope, intercept=intercept, color='red', linewidth=3)
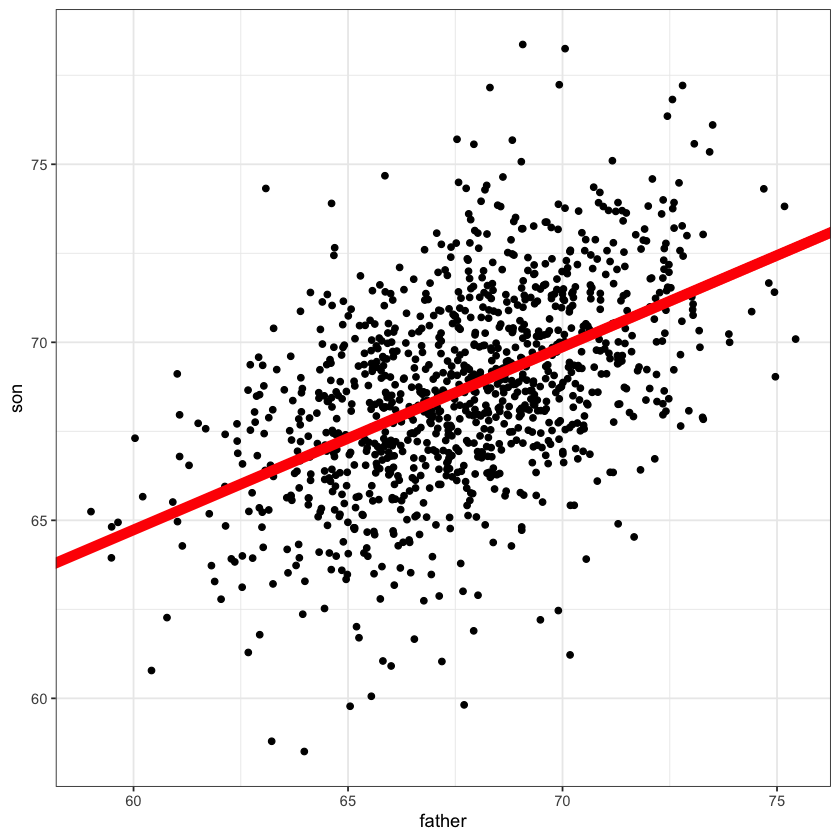
height.lm = lm(son ~ father, data=heights)
height.lm$coef
- (Intercept)
- 33.8866043540779
- father
- 0.514093038623309
What is a “regression” model?#
Model of the relationships between some covariates / predictors / features and an outcome.
A regression model is a model of the average outcome given the covariates.
Mathematical formulation#
For height data: a mathematical model:
\(f\) describes how mean of
sonvaries withfather\(\varepsilon\) is the random variation within the slice.
 {height=600 fig-align=”center”}
{height=600 fig-align=”center”}
Linear regression models#
A linear regression model says that the function \(f\) is a sum (linear combination) of functions of
father.Simple linear regression model:
Parameters of \(f\) are \((\beta_0, \beta_1)\)
Could also be a sum (linear combination) of fixed functions of
father:
Statistical model#
Symbol \(Y\) usually used for outcomes, \(X\) for covariates…
Model:
where \(\varepsilon_i \sim N(0, \sigma^2)\) are independent.
This specifies a distribution for the \(Y\)’s given the \(X\)’s, i.e. it is a statistical model.
 {height=600 fig-align=”center”}
{height=600 fig-align=”center”}
Regression equation#
The regression equation is our slicewise model.
Formally, this is a model of the conditional mean function
Book uses the notation \(\mu\{Y|X\}\).
Fitting the model#
We will be using least squares regression. This measures the goodness of fit of a line by the sum of squared errors, \(SSE\).
Least squares regression chooses the line that minimizes
In principle, we might measure goodness of fit differently:
\[ SAD(\beta_0, \beta_1) = \sum_{i=1}^n |Y_i - \beta_0 - \beta_1 \cdot X_i|.\]For some other loss function \(L\) we might try to minimize
\[ L(\beta_0,\beta_1) = \sum_{i=1}^n L(Y_i-\beta_0-\beta_1\cdot X_i) \]
Why least squares?#
With least squares, the minimizers have explicit formulae – not so important with today’s computer power.
Resulting formulae are linear in the outcome \(Y\). This is important for inferential reasons. For only predictive power, this is also not so important.
If assumptions are correct, then this is maximum likelihood estimation.
Statistical theory tells us the maximum likelihood estimators (MLEs) are generally good estimators.
Choice of loss function#
Suppose we try to minimize squared error over \(\mu\):
We know (by calculus) that the minimizer is the sample mean.
If we minimize absolute error over \(\mu\)
We know (similarly by calculus) that the minimizer(s) is (are) the sample median(s).
Visualizing the loss function#
Let’s take some a random scatter plot and view the loss function.
X = rnorm(50)
Y = 1.5 + 0.1 * X + rnorm(50) * 2
parameters = lm(Y ~ X)$coef
intercept = parameters[1]
slope = parameters[2]
D = data.frame(X, Y)
ggplot(D, aes(X, Y)) + geom_point() + geom_abline(slope=slope, intercept=intercept)
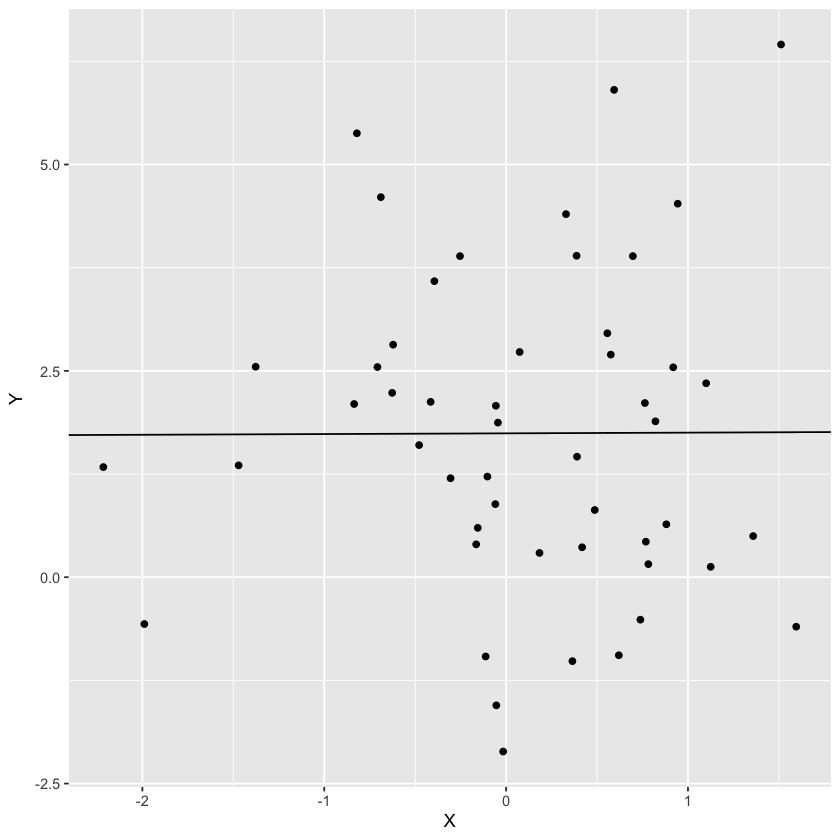
Let’s plot the loss as a function of the parameters. Note that the true intercept is 1.5 while the true slope is 0.1.
grid_intercept = seq(intercept - 2, intercept + 2, length=100)
grid_slope = seq(slope - 2, slope + 2, length=100)
loss_data = expand.grid(intercept=grid_intercept, slope=grid_slope)
loss_data$squared_error = numeric(nrow(loss_data))
for (i in 1:nrow(loss_data)) {
loss_data$squared_error[i] = sum((D$Y - D$X * loss_data$slope[i] - loss_data$intercept[i])^2)
}
squared_error_fig = (ggplot(loss_data, aes(intercept, slope, fill=squared_error)) +
geom_raster() +
scale_fill_gradientn(colours=c("gray","yellow","blue")))
squared_error_fig
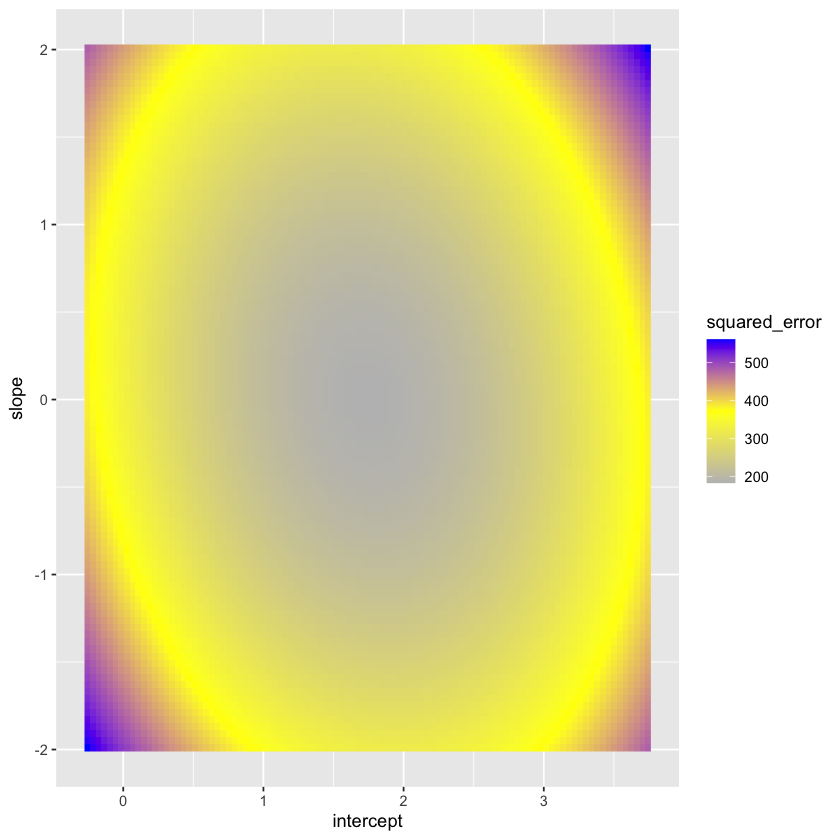
Let’s contrast this with the sum of absolute errors.
loss_data$absolute_error = numeric(nrow(loss_data))
for (i in 1:nrow(loss_data)) {
loss_data$absolute_error[i] = sum(abs(D$Y - D$X * loss_data$slope[i] - loss_data$intercept[i]))
}
absolute_error_fig = (ggplot(loss_data, aes(intercept, slope, fill=absolute_error)) +
geom_raster() +
scale_fill_gradientn(colours=c("gray","yellow","blue")))
absolute_error_fig
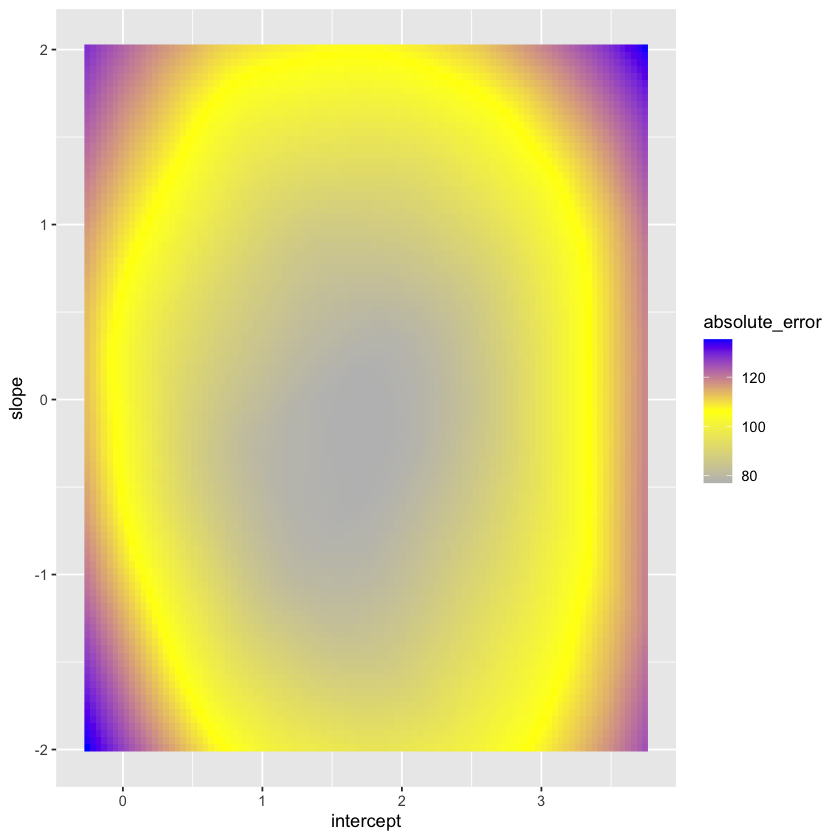
Geometry of least squares#
 {height=600 fig-align=”center”}
{height=600 fig-align=”center”}
Some things to note:
Minimizing sum of squares is the same as finding the point in the
X,1plane closest to \(Y\).The total dimension of the space is 1078.
The dimension of the plane is 2-dimensional.
The axis marked “\(\perp\)” should be thought of as \((n-2)\) dimensional, or, 1076 in this case.
Least squares#
 {height=600 fig-align=”center”}
{height=600 fig-align=”center”}
The (squared) lengths of the above vectors are important quantities in what follows.
Important lengths#
There are three to note:
An important summary of the fit is the ratio
Measures how much variability in \(Y\) is explained by \(X\).
Case study A: data suggesting the Big Bang#
url = 'https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/big_bang.csv'
big_bang = read.table(url, sep=',', header=TRUE)
with(big_bang, plot(Velocity, Distance, pch=23, bg='red'))
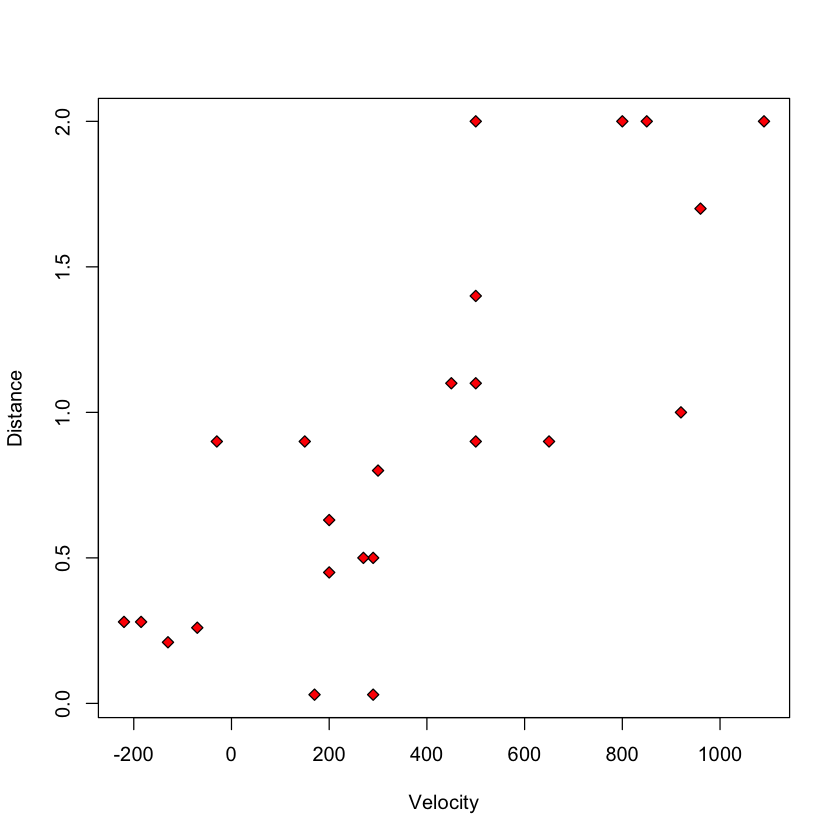
Let’s fit the linear regression model.
big_bang.lm = lm(Distance ~ Velocity, data=big_bang)
big_bang.lm
Call:
lm(formula = Distance ~ Velocity, data = big_bang)
Coefficients:
(Intercept) Velocity
0.399170 0.001372
Let’s look at the summary:
summary(big_bang.lm)
Call:
lm(formula = Distance ~ Velocity, data = big_bang)
Residuals:
Min 1Q Median 3Q Max
-0.76717 -0.23517 -0.01083 0.21081 0.91463
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.3991704 0.1186662 3.364 0.0028 **
Velocity 0.0013724 0.0002278 6.024 4.61e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.4056 on 22 degrees of freedom
Multiple R-squared: 0.6226, Adjusted R-squared: 0.6054
F-statistic: 36.29 on 1 and 22 DF, p-value: 4.608e-06
Hubble’s model#
Hubble’s theory of the Big Bang suggests that the correct slicewise (i.e. regression) model is
To fit without an intercept
hubble.lm = lm(Distance ~ Velocity - 1, data=big_bang)
summary(hubble.lm)
Call:
lm(formula = Distance ~ Velocity - 1, data = big_bang)
Residuals:
Min 1Q Median 3Q Max
-0.76767 -0.06909 0.22948 0.46056 1.03931
Coefficients:
Estimate Std. Error t value Pr(>|t|)
Velocity 0.0019214 0.0001913 10.04 7.05e-10 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.4882 on 23 degrees of freedom
Multiple R-squared: 0.8143, Adjusted R-squared: 0.8062
F-statistic: 100.9 on 1 and 23 DF, p-value: 7.05e-10
with(big_bang, plot(Velocity, Distance, pch=23, bg='red'))
slope = coef(big_bang.lm)[1]
intercept = coef(big_bang.lm)[2]
abline(slope, intercept, lwd=2)
abline(0, coef(hubble.lm)[1], col='blue', lwd=2)
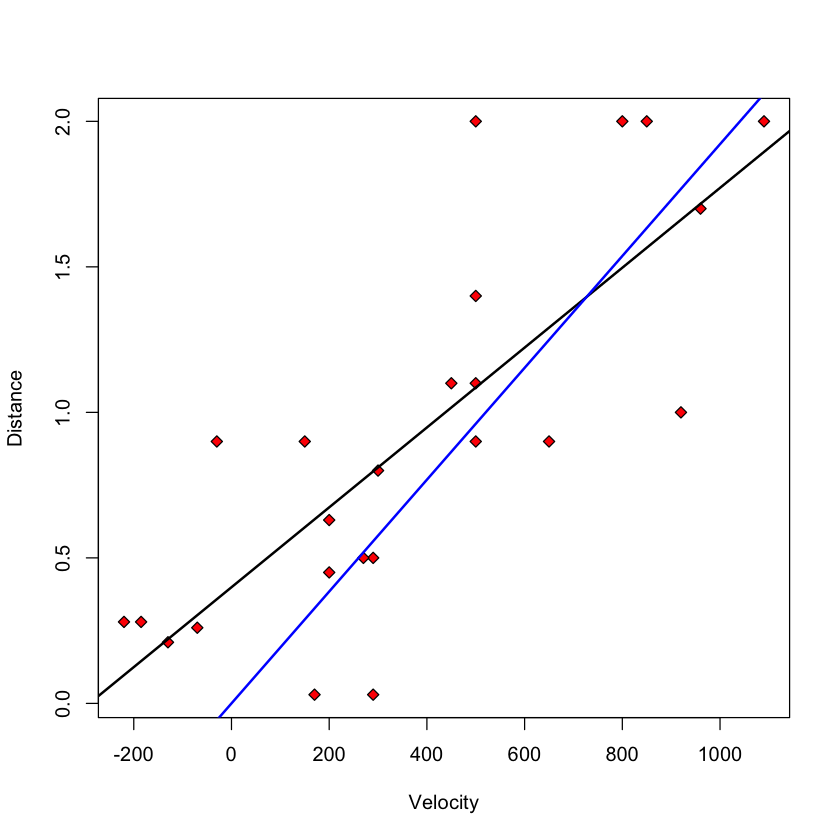
Least squares estimators#
There are explicit formulae for the least squares estimators, i.e. the minimizers of the error sum of squares.
For the slope, \(\hat{\beta}_1\), it can be shown that
Knowing the slope estimate, the intercept estimate can be found easily:
Example: big_bang#
beta.1.hat = with(big_bang, cov(Distance, Velocity) / var(Velocity))
beta.0.hat = with(big_bang, mean(Distance) - beta.1.hat * mean(Velocity))
c(beta.0.hat, beta.1.hat)
coef(big_bang.lm)
- 0.399170439725205
- 0.00137240753172474
- (Intercept)
- 0.399170439725205
- Velocity
- 0.00137240753172474
Estimate of \(\sigma^2\)#
The estimate most commonly used is
We’ll use the common practice of replacing the quantity \(SSE(\hat{\beta}_0,\hat{\beta}_1)\), i.e. the minimum of this function, with just \(SSE\).
The term MSE above refers to mean squared error: a sum of squares divided by its degrees of freedom. The degrees of freedom of SSE, the error sum of squares is therefore \(n-2\).
Mathematical aside#
We divide by \(n-2\) because some calculations tell us:
Above, the right hand side denotes a chi-squared distribution with \(n-2\) degrees of freedom.
Dividing by \(n-2\) gives an unbiased estimate of \(\sigma^2\) (assuming our modeling assumptions are correct).
Example: big_bang#
sigma_hat = sqrt(sum(resid(big_bang.lm)^2) / big_bang.lm$df.resid)
sigma_hat
summary(big_bang.lm)
Call:
lm(formula = Distance ~ Velocity, data = big_bang)
Residuals:
Min 1Q Median 3Q Max
-0.76717 -0.23517 -0.01083 0.21081 0.91463
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.3991704 0.1186662 3.364 0.0028 **
Velocity 0.0013724 0.0002278 6.024 4.61e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.4056 on 22 degrees of freedom
Multiple R-squared: 0.6226, Adjusted R-squared: 0.6054
F-statistic: 36.29 on 1 and 22 DF, p-value: 4.608e-06
Inference for the simple linear regression model#
 {height=400 fig-align=”center”}
{height=400 fig-align=”center”}
Remember: \(X\) can be fixed or random in our model…
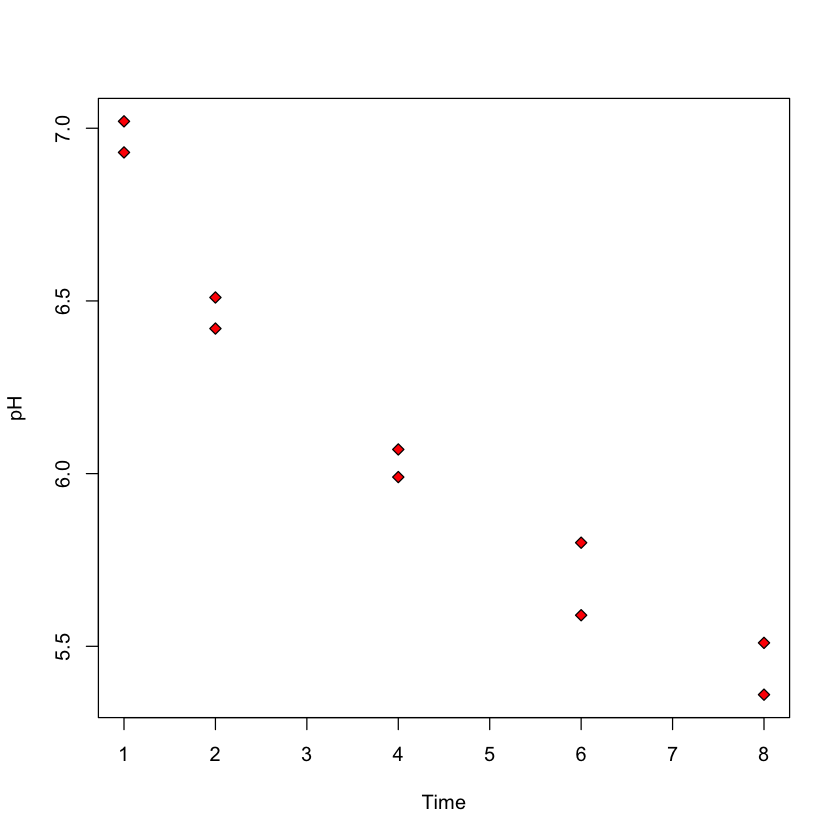
Case study B: predicting pH based on time after slaughter#
In this study, researches fixed \(X\) (
Time) before measuring \(Y\) (pH)Ultimate goal: how long after slaughter is
pHaround 6?
url = 'https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/meat_processing.csv'
meat_proc = read.table(url, sep=',', header=TRUE)
with(meat_proc, plot(Time, pH, pch=23, bg='red'))
Inference for \(\beta_0\) or \(\beta_1\)#
Recall our model
The errors \(\varepsilon_i\) are independent \(N(0, \sigma^2)\).
In our heights example, we might want to now if there really is a linear association between \({\tt son}=Y\) and \({\tt father}=X\). This can be answered with a hypothesis test of the null hypothesis \(H_0:\beta_1=0\). This assumes the model above is correct, AND \(\beta_1=0\).
Alternatively, we might want to have a range of values that we can be fairly certain \(\beta_1\) lies within. This is a confidence interval for \(\beta_1\).
A mathematical aside#
Let \(L\) be the subspace of \(\mathbb{R}^n\) spanned \(\pmb{1}=(1, \dots, 1)\) and \({X}=(X_1, \dots, X_n)\).
We can decompose \(Y\) as
In our model, \(\mu=\beta_0 \pmb{1} + \beta_1 {X} \in L\) so that
\[ \widehat{{Y}} = \mu + P_L{\varepsilon}, \qquad {e} = P_{L^{\perp}}{{Y}} = P_{L^{\perp}}{\varepsilon}\]Our assumption that \(\varepsilon_i\)’s are independent \(N(0,\sigma^2)\) tells us that: \({e}\) and \(\widehat{{Y}}\) are independent; \(\widehat{\sigma}^2 = \|{e}\|^2 / (n-2) \sim \sigma^2 \cdot \chi^2_{n-2} / (n-2)\).
Setup for inference#
All of this implies
Therefore,
The other quantity we need is the standard error or SE of \(\hat{\beta}_1\):
Testing \(H_0:\beta_1=\beta_1^0\)#
Suppose we want to test that \(\beta_1\) is some pre-specified value, \(\beta_1^0\) (this is often 0: i.e. is there a linear association)
Under \(H_0:\beta_1=\beta_1^0\)
\[\frac{\widehat{\beta}_1 - \beta^0_1}{\widehat{\sigma} \sqrt{\frac{1}{\sum_{i=1}^n(X_i-\overline{X})^2}}} = \frac{\widehat{\beta}_1 - \beta^0_1}{ \frac{\widehat{\sigma}}{\sigma}\cdot \sigma \sqrt{\frac{1}{ \sum_{i=1}^n(X_i-\overline{X})^2}}} \sim t_{n-2}.\]Reject \(H_0:\beta_1=\beta_1^0\) if \(|T| > t_{n-2, 1-\alpha/2}\).
Example: big_bang#
Let’s perform this test for the Big Bang data.
SE.beta.1.hat = with(big_bang, sigma_hat * sqrt(1 / sum((Velocity - mean(Velocity))^2)))
Tstat = (beta.1.hat - 0) / SE.beta.1.hat
c(estimate=beta.1.hat, SE=SE.beta.1.hat, T=Tstat)
summary(big_bang.lm)
- estimate
- 0.00137240753172474
- SE
- 0.00022782136747332
- T
- 6.0240509788243
Call:
lm(formula = Distance ~ Velocity, data = big_bang)
Residuals:
Min 1Q Median 3Q Max
-0.76717 -0.23517 -0.01083 0.21081 0.91463
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.3991704 0.1186662 3.364 0.0028 **
Velocity 0.0013724 0.0002278 6.024 4.61e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.4056 on 22 degrees of freedom
Multiple R-squared: 0.6226, Adjusted R-squared: 0.6054
F-statistic: 36.29 on 1 and 22 DF, p-value: 4.608e-06
We see that R performs our \(t\)-test in the second row of the
Coefficientstable.It is clear that
Distanceis correlated withVelocity.There seems to be some flaw in Hubble’s theory: we reject \(H_0:\beta_0=0\) at level 5%: \(p\)-value is 0.0028!
Why reject for large |T|?#
Logic is the same as other \(t\) tests: observing a large \(|T|\) is unlikely if \(\beta_1 = \beta_1^0\) (i.e. if \(H_0\) were true). \(\implies\) it is reasonable to conclude that \(H_0\) is false.
Common to report \(p\)-value:
2*(1 - pt(Tstat, big_bang.lm$df.resid))
summary(big_bang.lm)
Call:
lm(formula = Distance ~ Velocity, data = big_bang)
Residuals:
Min 1Q Median 3Q Max
-0.76717 -0.23517 -0.01083 0.21081 0.91463
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.3991704 0.1186662 3.364 0.0028 **
Velocity 0.0013724 0.0002278 6.024 4.61e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.4056 on 22 degrees of freedom
Multiple R-squared: 0.6226, Adjusted R-squared: 0.6054
F-statistic: 36.29 on 1 and 22 DF, p-value: 4.608e-06
Confidence interval for regression parameters#
Applying the above to the parameter \(\beta_1\) yields a confidence interval of the form
Earlier, we computed \(SE(\hat{\beta}_1)\) using this formula
\[ SE(a_0 \cdot \hat{\beta}_0 + a_1 \cdot \hat{\beta}_1) = \hat{\sigma} \sqrt{\frac{a_0^2}{n} + \frac{(a_0\cdot \overline{X} - a_1)^2}{\sum_{i=1}^n \left(X_i-\overline{X}\right)^2}}\]
with \((a_0,a_1) = (0, 1)\).
We also need to find the quantity \(t_{n-2,1-\alpha/2}\). This is defined by
In R, this is computed by the function qt.
alpha = 0.05
n = nrow(big_bang)
qt(1-0.5*alpha, n-2)
qnorm(1 - 0.5*alpha)
L = beta.1.hat - qt(0.975, big_bang.lm$df.resid) * SE.beta.1.hat
U = beta.1.hat + qt(0.975, big_bang.lm$df.resid) * SE.beta.1.hat
c(lower=L, upper=U)
- lower
- 0.000899934933428759
- upper
- 0.00184488013002073
We will not need to use these explicit formulae all the time, as R has some built in functions to compute confidence intervals.
confint(big_bang.lm)
confint(big_bang.lm, level=0.9)
| 2.5 % | 97.5 % | |
|---|---|---|
| (Intercept) | 0.1530719058 | 0.64526897 |
| Velocity | 0.0008999349 | 0.00184488 |
| 5 % | 95 % | |
|---|---|---|
| (Intercept) | 0.1954035267 | 0.60293735 |
| Velocity | 0.0009812054 | 0.00176361 |
Predicting the mean#
Once we have estimated a slope \((\hat{\beta}_1)\) and an intercept \((\hat{\beta}_0)\), we can predict the height of the son born to a father of any particular height by the plugging-in the height of the new father, \(F_{new}\) into our regression equation:
Confidence interval for the average height of sons born to a father of height \(F_{new}=70\) (or maybe \(65\)) inches:
predict(height.lm,
list(father=c(70, 65)),
interval='confidence',
level=0.90)
| fit | lwr | upr | |
|---|---|---|---|
| 1 | 69.87312 | 69.71333 | 70.03291 |
| 2 | 67.30265 | 67.13165 | 67.47366 |
new_fig = heights_fig + geom_abline(slope=slope, intercept=intercept, color='red', linewidth=3)
rect = data.frame(xmin=64.5, xmax=65.5, ymin=-Inf, ymax=Inf)
new_fig = new_fig + geom_rect(data=rect, aes(xmin=xmin, xmax=xmax, ymin=ymin, ymax=ymax),
color="grey20",
alpha=0.5,
inherit.aes = FALSE)
rect = data.frame(xmin=69.5, xmax=70.5, ymin=-Inf, ymax=Inf)
new_fig = new_fig + geom_rect(data=rect, aes(xmin=xmin, xmax=xmax, ymin=ymin, ymax=ymax),
color="grey20",
alpha=0.5,
inherit.aes = FALSE)
new_fig
Computing \(SE(\hat{\beta}_0 + 70 \cdot \hat{\beta}_1)\)#
We use the previous formula
\[ SE(a_0\cdot \hat{\beta}_0 + a_1\cdot\hat{\beta}_1) = \hat{\sigma} \sqrt{\frac{a_0^2}{n} + \frac{(a_0\cdot \overline{X} - a_1)^2}{\sum_{i=1}^n \left(X_i-\overline{X}\right)^2}}\]with \((a_0, a_1) = (1, 70)\).
Plugging in
As \(n\) grows (taking a larger sample), \(SE(\hat{\beta}_0 + 70 \hat{\beta}_1)\) should shrink to 0. Why?
Forecasting / prediction intervals#
Can we find an interval that covers the height of a particular son knowing only that her father’s height as 70 inches?
Must cover the variability of the new random variation \(\implies\) it must be at least as wide as \(\sigma\).
#| fig-align: center
rect = data.frame(xmin=69.5, xmax=70.5, ymin=-Inf, ymax=Inf)
heights_fig + geom_rect(data=rect, aes(xmin=xmin, xmax=xmax, ymin=ymin, ymax=ymax),
color="grey20",
alpha=0.5,
inherit.aes = FALSE)

selected_points = (father <= 70.5) & (father >= 69.5)
center_ = mean(son[selected_points])
sd_ = sd(son[selected_points])
L = center_ - qnorm(0.95) * sd_
U = center_ + qnorm(0.95) * sd_
c(estimate=center_, lower=L, upper=U)
- estimate
- 69.7684482608696
- lower
- 65.6735353469882
- upper
- 73.8633611747509
With so much data in our heights example, this 90% interval will have width roughly \(2 \cdot 1.96 \cdot \hat{\sigma}\)
sigma_hat_height = sqrt(sum(resid(height.lm)^2) / height.lm$df.resid)
c(2 * qnorm(0.95) * sigma_hat_height, U-L)
- 8.01555526222726
- 8.18982582776275
Formula#
Actual width will depend on how accurately we have estimated \((\beta_0, \beta_1)\) as well as \(\hat{\sigma}\).
The final interval is
Computed in
Ras follows
predict(height.lm, list(father=70), interval='prediction', level=0.90)
| fit | lwr | upr | |
|---|---|---|---|
| 1 | 69.87312 | 65.8587 | 73.88753 |