Assumptions for t tests#
Download#
Outline#
Case studies:
Cloud seeding
Effects of agent orange
Robustness and resistance of two-sample \(t\)-tests
Transformations
require(ggplot2)
set.seed(0)
Loading required package: ggplot2
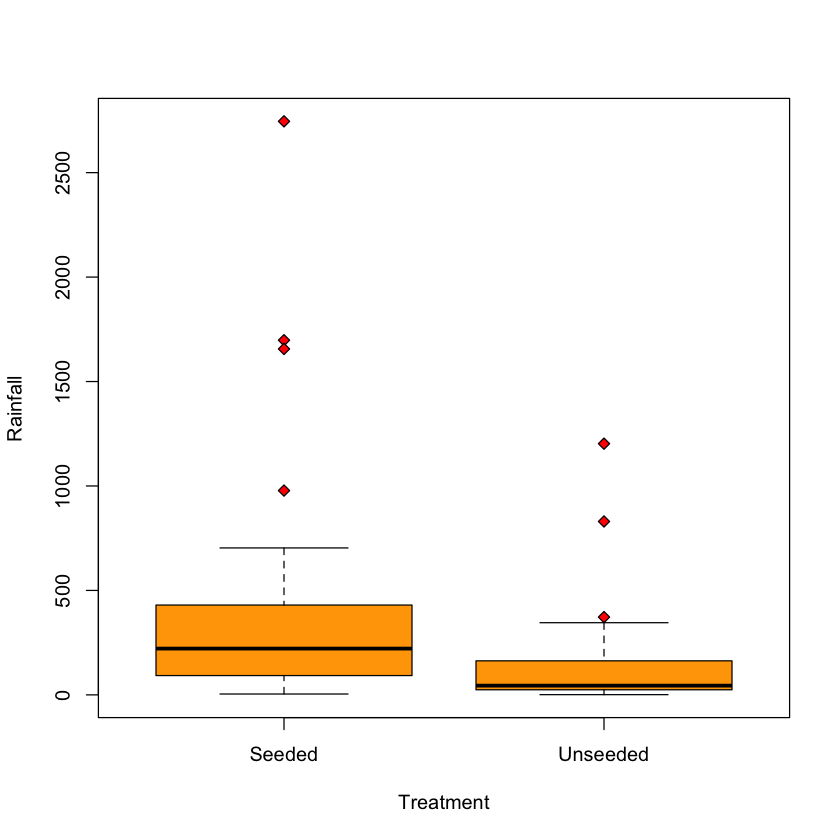
Case study A: effect of cloud seeding#
rainfall = read.csv('https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/rainfall.csv', header=TRUE)
boxplot(Rainfall ~ Treatment,
data=rainfall,
col='orange',
pch=23,
bg='red')
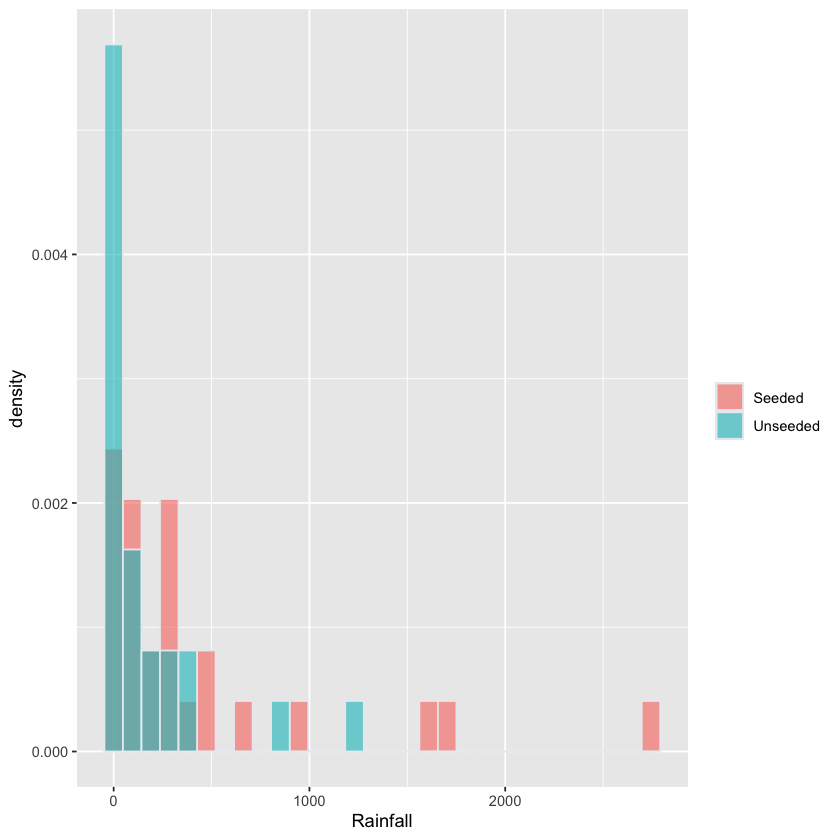
Histogram of Rainfall stratified by Treatment#
# this plot is for visualization only
# students not expected to reproduce
rainfall = read.csv('https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/rainfall.csv', header=TRUE)
fig <- (ggplot(rainfall, aes(x=Rainfall, fill=Treatment)) +
geom_histogram(aes(y=after_stat(density)),
color="#e9ecef",
alpha=0.6,
position='identity',
bins=30) +
labs(fill=""))
fig
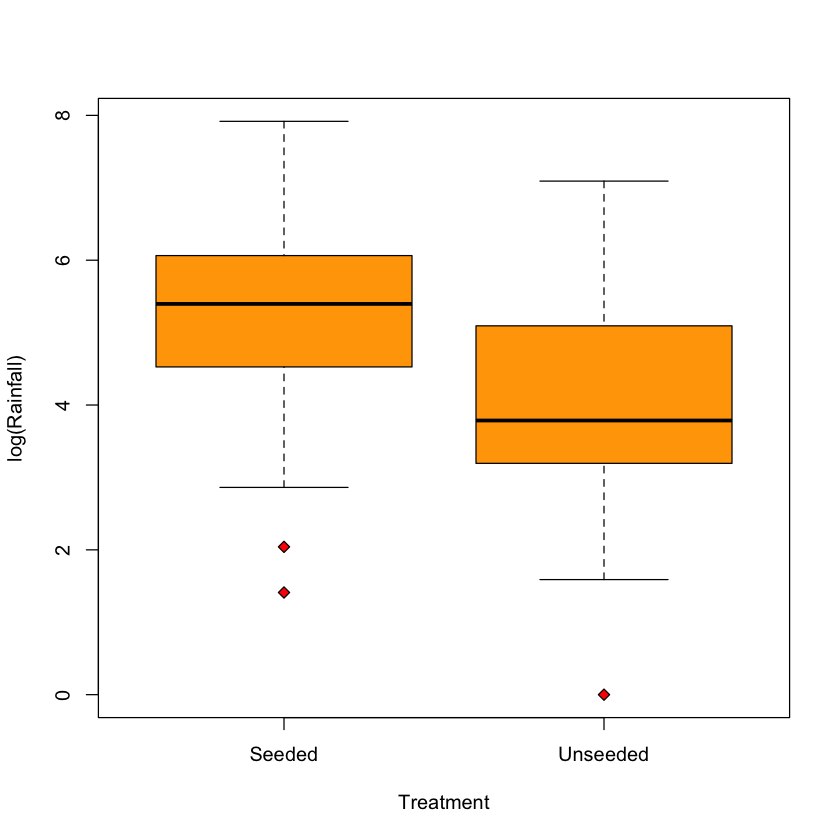
Practical tip: log transformation#
boxplot(log(Rainfall) ~ Treatment,
data=rainfall,
col='orange',
pch=23,
bg='red')
Histogram of log(Rainfall) stratified by Treatment#
# this plot is for visualization only
# students not expected to reproduce
rainfall$logRainfall = log(rainfall$Rainfall)
fig <- (ggplot(rainfall, aes(x=logRainfall, fill=Treatment)) +
geom_histogram(aes(y=after_stat(density)),
color="#e9ecef",
alpha=0.6,
position='identity',
bins=30) +
labs(fill=""))
fig
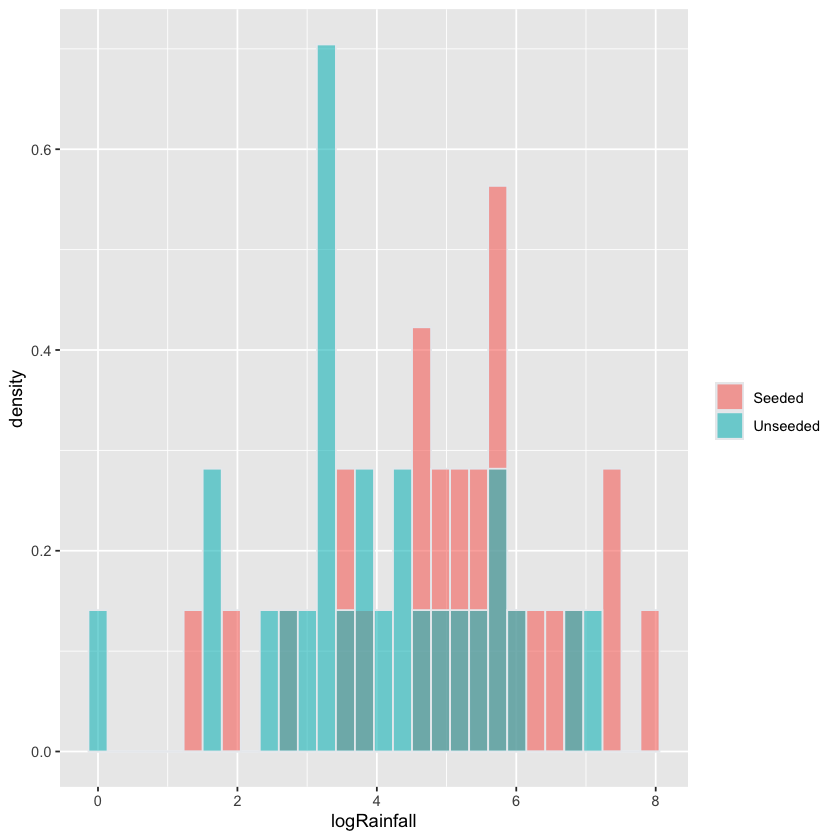
Does cloud seeding help?#
Histogram on log scale has similar shape for both groups \(\implies\) \(t\)-test probably well founded here.
t.test(log(Rainfall) ~ Treatment,
var.equal=TRUE,
data=rainfall)
Two Sample t-test
data: log(Rainfall) by Treatment
t = 2.5444, df = 50, p-value = 0.01408
alternative hypothesis: true difference in means between group Seeded and group Unseeded is not equal to 0
95 percent confidence interval:
0.240865 2.046697
sample estimates:
mean in group Seeded mean in group Unseeded
5.134187 3.990406
Robustness of two sample \(t\)-tests#
Our analysis of
beakspresumed \(\sigma^2_A=\sigma^2_B\) (as well as normality)What happens if:
Unequal variance: \(\sigma^2_A \neq \sigma^2_B\)?
Populations are not normal?
Observations are not independent?
Data are contaminated with outliers?
Mental model#
 {width=600 fig-align=”center”}
{width=600 fig-align=”center”}
Draw \(n_A\) samples from orange, \(n_B\) samples from purple.
Non-normality#
Equal sample size \(n_A \approx n_B\)#
Some effect of long tails and skewness
Unequal sample size \(n_A \neq n_B\)#
Substantially affected by skewness
Skewness#
If skewness of distributions is quite different, \(t\) tools are affected for small and moderate sample sizes.
Unequal standard deviations \(\sigma^2_A \neq \sigma^2_B\)#
If \(n_A \approx n_B\) then small effect.
Larger issue if \(n_A \neq n_B\).
Observations not being independent#
\(t\)-tests work poorly here
Main problem is that \(SE\) will be off, usually we underestimate it…
Outlier#
A point in the data that is far from the others.
Could be an accident in dataset construction, or could be due to long tails…
Try analyzing data with / without candidate outliers
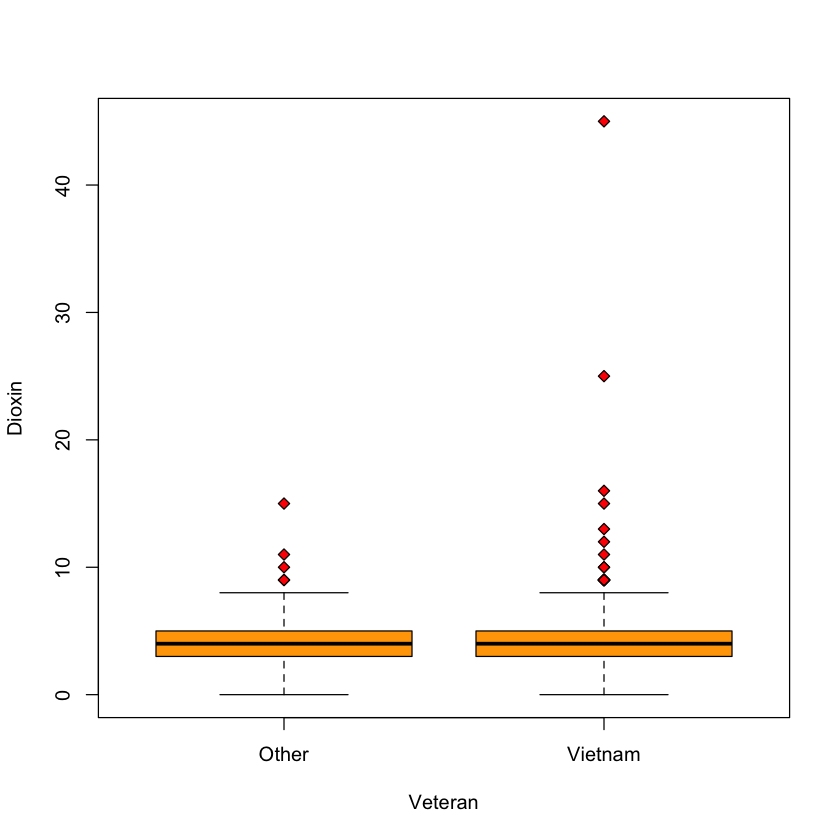
Case study B: dioxin in veterans#
agent_orange = read.csv('https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/agent_orange.csv', header=TRUE)
boxplot(Dioxin ~ Veteran,
data=agent_orange,
col='orange',
pch=23,
bg='red')
Histogram of Dioxin stratified by Veteran#
# this plot is for visualization only
# students not expected to reproduce
agent_orange = read.csv('https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/agent_orange.csv', header=TRUE)
fig <- (ggplot(agent_orange, aes(x=Dioxin, fill=Veteran)) +
geom_histogram(aes(y=after_stat(density)),
color="#e9ecef",
alpha=0.6,
position='identity',
bins=30) +
labs(fill=""))
fig
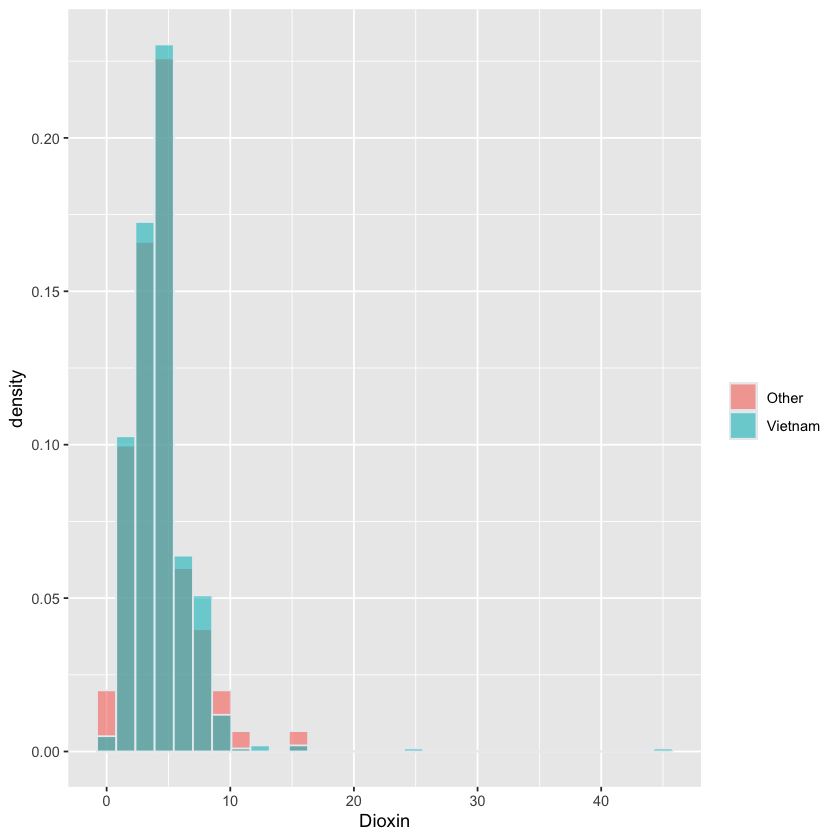
Outliers?#
Two Vietnam vets with level > 20
Histograms have similar shape, so skewness similar + large sample sizes \(\implies\) \(t\)-test probably not too bad.
t.test(Dioxin ~ Veteran,
var.equal=TRUE,
data=agent_orange)
Two Sample t-test
data: Dioxin by Veteran
t = -0.26302, df = 741, p-value = 0.7926
alternative hypothesis: true difference in means between group Other and group Vietnam is not equal to 0
95 percent confidence interval:
-0.6305128 0.4815229
sample estimates:
mean in group Other mean in group Vietnam
4.185567 4.260062
agent_orange$keep = agent_orange$Dioxin < 20
t.test(Dioxin ~ Veteran,
var.equal=TRUE,
subset=keep,
data=agent_orange)$stat
Transformations#
We saw earlier that histogram for
log(Rainfall)looked more “normal”.Using \(t\)-test on
log(Rainfall)has \(\mu_{\tt Treated}\) as the mean of the log of rainfall after seeding…
Parameter \(\mu_{\tt Treated} - \mu_{\tt Untreated}\)#
Acts multiplicatively
Interpretation#
As noted in the book, the estimated effect is on log scale.
Can be interpreted reasonably well when distribution of log-transformed data are symmetric.
We estimate
Treatedhas \(e^{5.13-3.99}\) multiplicative effect onmedian(Rainfall).