ANOVA#
Download#
Outline#
ANOVA models: variables with only categorical variables
Case studies:
A. Seaweed regeneration time based on grazers
B. Pygmalion effect
Case study A: Seaweed grazers#
url = 'https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/seaweed_grazers.csv'
seaweed.df = read.csv(url, sep=',', header=TRUE)
head(seaweed.df)
| Cover | Block | Treat | |
|---|---|---|---|
| <int> | <chr> | <chr> | |
| 1 | 14 | B1 | C |
| 2 | 23 | B1 | C |
| 3 | 22 | B2 | C |
| 4 | 35 | B2 | C |
| 5 | 67 | B3 | C |
| 6 | 82 | B3 | C |
Treat: which animals were allowed into grazeBlock: which of 8 blocks measuredCover: what percentage of each block had seaweed covered at time of measurement
Case study B: Pygmalion effect#
url = 'https://raw.githubusercontent.com/StanfordStatistics/stats191-data/main/Sleuth3/pygmalion.csv'
pygmalion.df = read.csv(url, sep=',', header=TRUE)
head(pygmalion.df)
| Company | Treat | Score | |
|---|---|---|---|
| <chr> | <chr> | <dbl> | |
| 1 | C1 | Pygmalion | 80.0 |
| 2 | C1 | Control | 63.2 |
| 3 | C1 | Control | 69.2 |
| 4 | C2 | Pygmalion | 83.9 |
| 5 | C2 | Control | 63.1 |
| 6 | C2 | Control | 81.5 |
Company: ID of companyTreat: whetherPygmalionorControlScore: outcome of interest
Additive model#
pygm_add.lm = lm(Score ~ Treat + Company, data=pygmalion.df)
anova(pygm_add.lm)
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | <dbl> | |
| Treat | 1 | 327.3410 | 327.34102 | 7.568540 | 0.01314152 |
| Company | 9 | 682.5171 | 75.83524 | 1.753407 | 0.14844454 |
| Residuals | 18 | 778.5039 | 43.25022 | NA | NA |
Saturated model#
pygm_sat.lm = lm(Score ~ Treat + Company + Treat:Company, data=pygmalion.df)
anova(pygm_sat.lm)
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | <dbl> | |
| Treat | 1 | 327.3410 | 327.34102 | 6.3079589 | 0.03322533 |
| Company | 9 | 682.5171 | 75.83524 | 1.4613676 | 0.29050766 |
| Treat:Company | 9 | 311.4639 | 34.60710 | 0.6668892 | 0.72211560 |
| Residuals | 9 | 467.0400 | 51.89333 | NA | NA |
Graphical check of additivity#
with(pygmalion.df,
interaction.plot(Company, Treat, Score,
type='p',
col=c('red', 'blue'),
lwd=2))

Two-way ANOVA model#
Generalization of \(t\)-test: more than one grouping variable.
Two-way ANOVA model:
\(r=8\) groups in first factor (
Block)\(m=6\) groups in second factor (
Treat)\(n_{ij}\) in each combination of factor variables.
Model: $\(Y_{ijk} = \mu + \alpha_i + \beta_j + (\alpha \beta)_{ij} + \varepsilon_{ijk} , \qquad \varepsilon_{ijk} \sim N(0, \sigma^2).\)$
In
seaweed.df, \(r=8\), \(m=6\), \(n_{ij}=2\) for all \((i,j)\).
Parameterization#
Some constraints are needed, again for identifiability. Let’s not worry too much about the details…
Constraints:
\(\sum_{i=1}^r \alpha_i = 0\)
\(\sum_{j=1}^m \beta_j = 0\)
\(\sum_{j=1}^m (\alpha\beta)_{ij} = 0, 1 \leq i \leq r\)
\(\sum_{i=1}^r (\alpha\beta)_{ij} = 0, 1 \leq j \leq m.\)
ANOVA table#
Source |
df |
E(MS) |
|---|---|---|
A |
r-1 |
\(\sigma^2 + nm\frac{\sum_{i=1}^r \alpha_i^2}{r-1}\) |
B |
m-1 |
\(\sigma^2 + nr\frac{\sum_{j=1}^m \beta_j^2}{m-1}\) |
A:B |
(m-1)(r-1) |
\(\sigma^2 + n\frac{\sum_{i=1}^r\sum_{j=1}^m (\alpha\beta)_{ij}^2}{(r-1)(m-1)}\) |
Error |
(n-1)mr |
\(\sigma^2\) |
Tests using the ANOVA table#
Rows of the ANOVA table can be used to test various of the hypotheses we started out with.
Under \(H_0:\) no interactions
Test of additivity#
\(H_0\):
pygm_add.lmis the true model
anova(pygm_add.lm, pygm_sat.lm)
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 18 | 778.5039 | NA | NA | NA | NA |
| 2 | 9 | 467.0400 | 9 | 311.4639 | 0.6668892 | 0.7221156 |
\(F\)-statistic \(\approx 0.67\) – very little evidence against \(H_0\).
Note: this \(F\)-statistic also appears in
anova(pygm_sat.lm).
Case study A: seaweed regeneration#
seaweed.df$Block = factor(seaweed.df$Block, levels=c('B1','B2','B7','B5', 'B8', 'B6', 'B3', 'B4'))
Before calling
interaction.plotwe’ll orderBlockas book does for Fig 13.9: ordered by total response.
Interaction plot for seaweed#
with(seaweed.df,
interaction.plot(Block, Treat, Cover,
type='b',
col=c('red', 'blue'),
lwd=2))
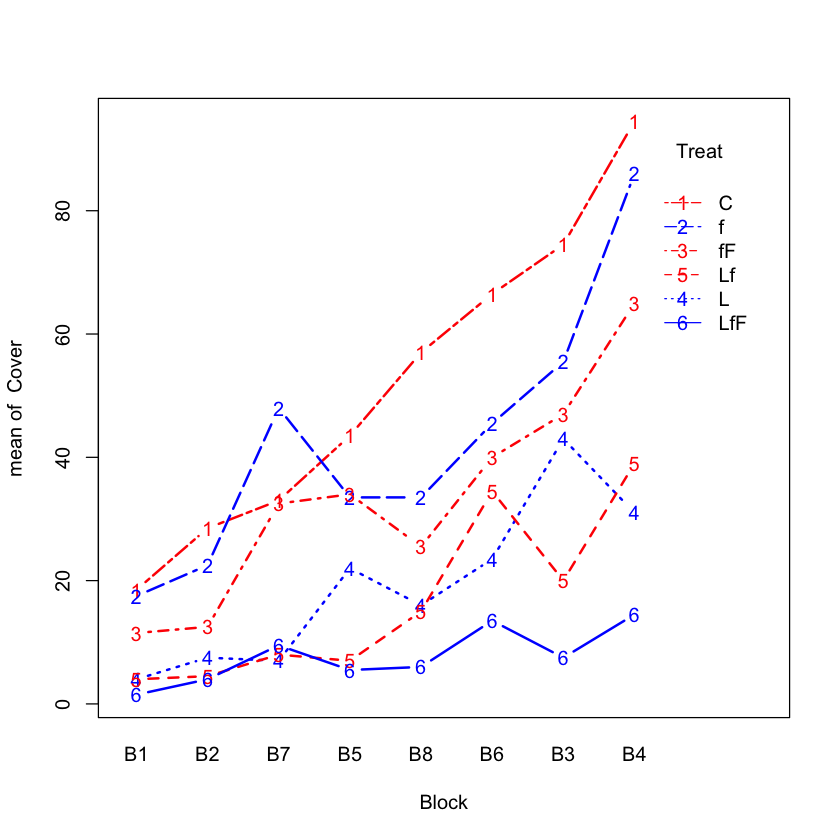
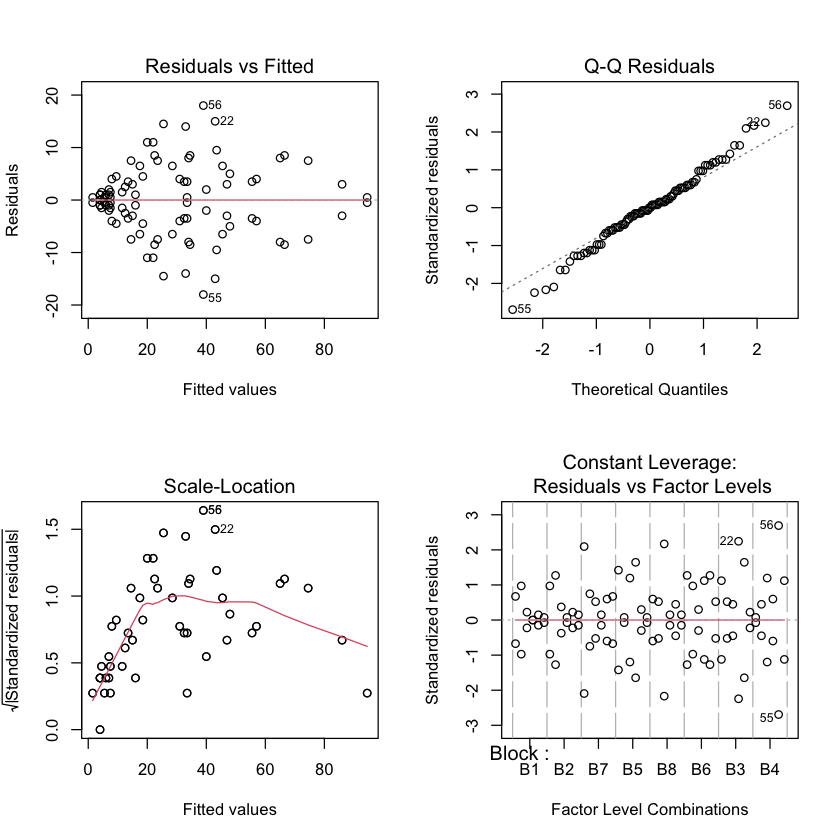
No transformation#
seaweed_sat.lm = lm(Cover ~ Block + Treat + Treat:Block, data=seaweed.df)
par(mfrow=c(2,2))
plot(seaweed_sat.lm)
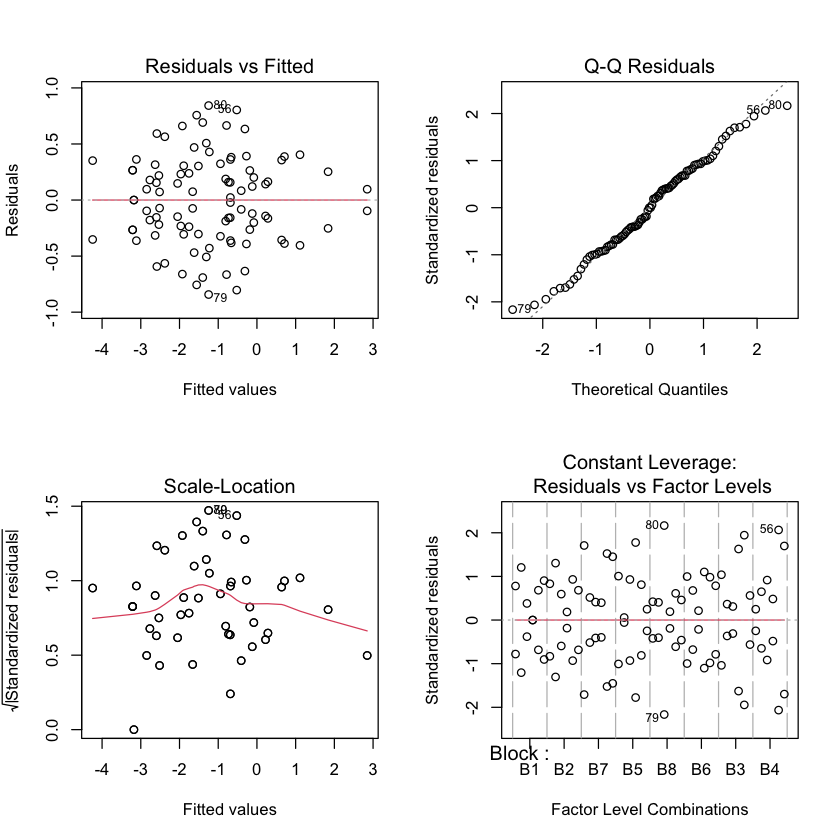
logit transformed#
seaweed.df$lCover = with(seaweed.df, log(Cover / (100 - Cover)))
seaweed_lsat.lm = lm(lCover ~ Block + Treat + Treat:Block, data=seaweed.df)
par(mfrow=c(2,2))
plot(seaweed_lsat.lm)
Interaction plot for logit transformed data#
with(seaweed.df,
interaction.plot(Block, Treat, lCover,
type='b',
col=c('red', 'blue'),
lwd=2))
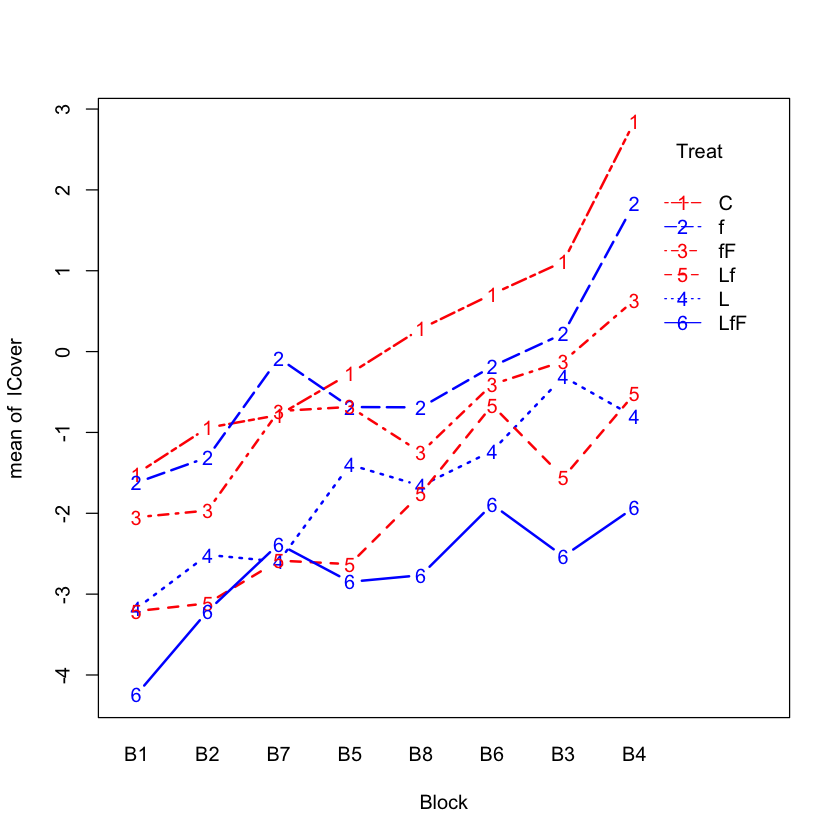
ANOVA table#
anova(seaweed_lsat.lm)
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | <dbl> | |
| Block | 7 | 76.23861 | 10.8912304 | 35.963371 | 5.390313e-17 |
| Treat | 5 | 96.99322 | 19.3986445 | 64.055265 | 4.510728e-20 |
| Block:Treat | 35 | 15.23041 | 0.4351546 | 1.436902 | 1.209129e-01 |
| Residuals | 48 | 14.53643 | 0.3028423 | NA | NA |
Another test for Treat#
seaweed_ladd.lm = lm(lCover ~ Block + Treat, data=seaweed.df)
anova(seaweed_ladd.lm)
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | <dbl> | |
| Block | 7 | 76.23861 | 10.8912304 | 30.36843 | 2.133837e-20 |
| Treat | 5 | 96.99322 | 19.3986445 | 54.08997 | 1.094513e-24 |
| Residuals | 83 | 29.76684 | 0.3586366 | NA | NA |
Difference between the two \(F\)-statistics: estimate of \(\sigma^2\)
Inference about specific linear combinations#
Do large fish have an effect on the regeneration ratio? If so, how much? (see Display 13.13)#
linear_comb = 0 * coef(seaweed_ladd.lm)
linear_comb['TreatLfF'] = 0.5
linear_comb['TreatfF'] = 0.5
linear_comb['TreatLf'] = -0.5
linear_comb['Treatf'] = -0.5
estimate = sum(linear_comb * coef(seaweed_ladd.lm))
Standard error#
var_est = sum(linear_comb * (vcov(seaweed_ladd.lm) %*% linear_comb))
SE = sqrt(var_est)
T_stat = estimate / SE
pval = 2 * pt(abs(T_stat), seaweed_ladd.lm$df.resid, lower=FALSE)
c(estimate=estimate, T_stat=T_stat, SE=SE, pval=pval)
- estimate
- -0.614025727666025
- T_stat
- -4.10127815915699
- SE
- 0.149715699310733
- pval
- 9.53986852987613e-05
Some caveats about R formulae#
While we see that it is straightforward to form the
interactions test using our usual anova function approach, we generally
cannot test for main effects by this approach.
lm1 = lm(lCover ~ Treat + Block:Treat, data=seaweed.df)
anova(lm1, seaweed_lsat.lm)
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 48 | 14.53643 | NA | NA | NA | NA |
| 2 | 48 | 14.53643 | 0 | -1.065814e-14 | NA | NA |
sum((resid(lm1) - resid(seaweed_lsat.lm))^2)